Biology:Eremothecium gossypii
| Eremothecium gossypii | |
|---|---|
| |
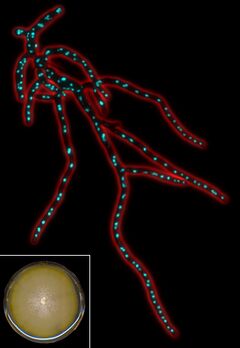
| Fluorescent micrograph of Template:Btname mycelium. | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Subphylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | E. gossypii
|
| Subspecies: | ATCC 10895, FDAG
|
| Binomial name | |
| Eremothecium gossypii (S.F. Ashby & W. Nowell) Kurtzman, 1995
| |
| Synonyms | |
Template:Btname[1] (also known as Ashbya gossypii[2]) is a filamentous fungus or mold closely related to yeast, but growing exclusively in a filamentous way. It was originally isolated from cotton as a pathogen causing stigmatomycosis by Ashby and Nowell in 1926. This disease affects the development of hair cells in cotton bolls and can be transmitted to citrus fruits, which thereupon dry out and collapse (dry rot disease). In the first part of the 20th century, Template:Btname and two other fungi causing stigmatomycosis (Template:Btname, Template:Btname) made it virtually impossible to grow cotton in certain regions of the subtropics, causing severe economical losses. Control of the spore-transmitting insects - cotton stainer (Template:Btname) and Template:Btname (antestia bugs) - permitted full eradication of infections. Template:Btname was recognized as a natural overproducer of riboflavin (vitamin B2), which protects its spores against ultraviolet light. This made it an interesting organism for industries, where genetically modified strains are still used to produce this vitamin.
Template:Btname as a model organism
A few years ago, Template:Btname became recognized as an attractive model to study the growth of long and multinucleate fungal cells (hyphae) because of its small genome, haploid nuclei, and efficient gene targeting methods. It is generally assumed that a better understanding of filamentous fungal growth will greatly stimulate the development of novel fungicides. Its use as a model organism is particularly promising because of the high level of gene order conservation (synteny) between the genomes of Template:Btname and the yeast Template:Btname.[citation needed]
Genome
The complete sequencing and annotation of the entire Template:Btname genome, as published in 2004, was initiated when a significant degree of gene synteny was observed in preliminary studies in comparison to the genome of budding yeast, Template:Btname. This not only helped to improve gene annotation of Template:Btname, but also allowed the reconstruction of the evolutionary history of both organisms. Template:Btname and Template:Btname originated from a common ancestor which carried about 5000 genes. Divergence of these two close relatives started some 100 million years ago. One branch of evolution involving up to 100 viable genome rearrangements (translocations and inversions), a few million base pair changes, and a limited number of gene deletions, duplications and additions lead to modern Template:Btname with its 4718 protein-coding genes and 9.2 million base pairs (smallest genome of a free-living eukaryote yet characterized) spread over seven chromosomes. The genome of Template:Btname underwent a more eventful evolution, which includes a whole-genome duplication.[citation needed]
Despite the long evolutionary history of the two organisms and fundamentally different ways of growth and development, the complete synteny map of both genomes reveals 95% of Template:Btname genes are orthologs of Template:Btname genes, and 90% map within blocks of synteny (syntenic homologs).
Growth, development and morphology
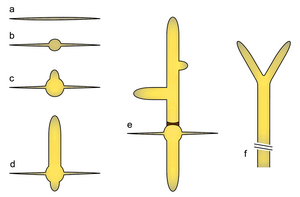
a) Ungerminated spore
b) Isotropic growth phase in the germ bubble
c) Unipolar germling
d) Emergence of a second germ tube
e) Emergence of lateral branches and septum generation
f) Apical branching in mature hypha
The Template:Btname life cycle starts with the only known phase of isotropic growth in wild type: germination of the haploid spore to form a germ bubble. This is followed by apical growth, extending two germ tubes in succession on opposing sites of the germ bubble. More axes of polarity are established with lateral branch formation in young mycelium. Maturation is characterized by apical branching (tip splitting) and a dramatic increase of growth speed (up to 200 μm/h at 30 °C), which enables it to cover an 8 cm Petri dish of full medium in about seven days. Sporulation is thought to be induced by nutrient deprivation, leading to contraction at the septa, cytokinesis and subsequent abscission of sporangia which contain up to eight haploid spores. Hyphae are compartmentalized by septa, which in young parts appear as rings that allow transfer of nuclei and in older parts may appear as closed discs. Compartments typically contain around eight nuclei.
References
- ↑ Gastmann, Selina; Dünkler, Alexander; Walther, Andrea; Klein, Keith; Wendland, Jürgen (2007). "A molecular toolbox for manipulating Eremothecium coryli". Microbiological Research 162 (4): 299–307. doi:10.1016/j.micres.2007.05.008. PMID 17716882.
- ↑ "Species Fungorum - GSD Species". http://www.speciesfungorum.org/Names/GSDSpecies.asp?RecordID=434470.
External links
- Ashby S. F.; Nowell W. (1926). "The Fungi of Stigmatomycosis.". Ann Bot os-40 (1): 69–84. doi:10.1093/oxfordjournals.aob.a090018.
- Dietrich F. S.; Voegeli S.; Brachat S.; Lerch A.; Gates K.; Steiner S.; Mohr C.; Pohlmann R. et al. (2004). "The Template:Btname genome as a tool for mapping the ancient Template:Btname genome.". Science 304 (5668): 304–307. doi:10.1126/science.1095781. PMID 15001715.
- Philippsen P.; Kaufmann A.; Schmitz H. P. (2005). "Homologues of yeast polarity genes control the development of multinucleated hyphae in Template:Btname.". Current Opinion in Microbiology 8 (4): 370–377. doi:10.1016/j.mib.2005.06.021. PMID 16023404.
- Gladfelter A. S. (2006). "Nuclear anarchy: asynchronous mitosis in multinucleated fungal hyphae.". Current Opinion in Microbiology 9 (6): 547–552. doi:10.1016/j.mib.2006.09.002. PMID 17045513.
- Kaufman A. (2008). Polarized growth and septation in the filamentous ascomycete Template:Btname analyzed by live cell imaging.
- "The Template:Btname Genome Database". http://agd.vital-it.ch/index.html.
- "Peter Philippsen's lab at the Biozentrum in Basel, Switzerland". http://www.biozentrum.unibas.ch/philippsen/index.html.
- "Hans-Peter Schmitz' lab at Universität Osnabrück, Germany". Archived from the original on 2007-03-14. https://web.archive.org/web/20070314063855/http://www.biologie.uni-osnabrueck.de/genetik/index.php?menuid=12.
- "Amy Galdfelter's lab at University of North Carolina - Chapel Hill, USA". https://gladfelterlab.web.unc.edu/.
- "Jürgen Wendland's lab at the CRC in Kopenhagen, Denmark". 29 September 2023. http://www.hki-jena.de/index.php/219e282223f034e143acfb8444d2ab6c/2/171.
- "Fred Dietrich's lab at Duke University, USA". Archived from the original on 2007-03-18. https://web.archive.org/web/20070318175338/http://labs.genome.duke.edu/DietrichLab/.
Wikidata ☰ Q27061531 entry
 |

