Biology:C1orf68
Chromosome 1 open reading frame 68, or C1orf68, is a human gene which encodes for skin-specific protein 32. C1orf68 gene is expressed in the skin,[1] is a part of the epidermal differentiation complex, and potentially plays a role in epidermal cornification, and epidermal barrier function.[2][3]
Gene
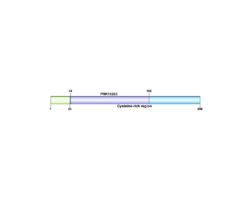
C1orf68 is mapped on the plus strand of chromosome 1 at 1q21.3, that spans 949 base pairs in the human genome.[4][5] Other aliases include Late envelope protein 7 (LEP7), XP32, Skin-Specific Protein (Xp32).[4] This gene has only 1 exon, and no introns. It is a part of the epidermal differentiation complex (1q21).
Protein
Skin-specific protein 32 has only one isoform, and has a sequence length of 250 amino acids.[6] It has a molecular mass of 26 kDa,[7][8] and a predicted pI value of 8.41.[8] It was noted that the amino acid sequence contained high levels of cysteine relative to other human protein sequences.[9]
Domains, repeats
Skin-specific protein 32 has one domain, PRK10264, which is a DNA translocase FtsK.[4] It also contains a cysteine rich region, which is shown to be conserved across most mammal orthologs, excluding Monotremes.[10]
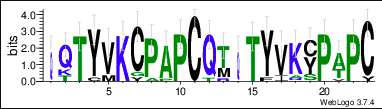
The protein sequence also contains a repeat sequence, the three continuous repeat sequences are located from amino acid position Gln65 to Cys127.[11] The repeat sequences can be observed in the conceptual translation on the right. They are within the DNA translocase Ftsk domain and the cysteine rich region. The repeat sequences are conserved across mammal orthologs. The conservation of each individual amino acid can be observed in the LOGO below.
Gene level regulation
Promoter
One promoter was identified for C1orf68 using ElDorado Genomatics.[12] This promoter, GXP_1818199, spans 1,040 bases and overlaps C1orf68 by 40 bases.[12] Since C1orf68 does not contain a 5'-UTR, the promoter overlaps the start codon, which can be visualized in the diagram below.
Expression pattern
C1orf68 is expressed in a select few tissues, specifically in the skin and in breast tissue.[13] In humans, C1orf68 protein abundance is moderate.[14] In terms of specific cell types within the skin, C1orf68 is expressed in suprabasal keratinocytes, which are a type of epithelial cell.[15] It has also been noted that C1orf68 is moderately expressed in stratum corneum and granular layer of skin.[16] This could be because the protein remains in the cell as it differentiates and matures.
Transcript level regulation
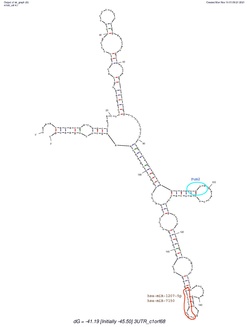
C1orf68 does not contain a 5'-UTR, but does contain a 3'-UTR. The predicted secondary structure of C1orf68's 3'-UTR mRNA contains various stem loops. The stem loop containing PUM2 RNA protein binding site, which was shown in all of the predicted structures created by mFold.[17]
Protein level regulation
Subcellular localization
Skin-specific protein 32 is predicted to be localized in the cytoplasm. The protein has been shown to occupy the cytoplasm within skin cells, which can be observed in the immunofluorescence staining in Human Protein Atlas, Subcellular.[18]
Evolution
Paralogs
There are no known paralogs of C1orf68.
Orthologs
C1orf68 has a range of orthologs within mammals, and some amphibians, specifically shown in two frog species. The ortholog sequence similarity percentages range from 96 to 23%. There are no orthologs in birds, fish, and reptiles but there was a few in amphibians. Additionally, within the mammals, there was no orthologs in Cetacea (marine mammals).[19] The most highly conserved amino acids across mammals and amphibians with available sequences are Pro61, Pro73, Pro126, Pro182, which are all proline amino acids.
| Genus Species | Common Name | Taxonomic Group | Divergence Data (MYA) Median Time[20] | Accession Number | Query Cover | Sequence Length (aa) | Sequence Identity (%) | Sequence Similarity (%) |
| Homo sapiens | Human | Primates | 0 | NP_001019850 | 100% | 250 | 100% | 100% |
| Rhinopithecus roxellana | Golden snub-nosed monkey | Primates | 29 | XP_030792113 | 98% | 250 | 94% | 96% |
| Callithrix jacchus | Common marmoset | Primates | 43 | XP_035135776 | 96% | 256 | 83% | 85% |
| Cavia porcellus | Guinea pig | Rotentia | 89 | XP_005007858 | 96% | 249 | 80% | 84% |
| Ochotona curzoniae | Plateau pika | Glires | 89 | XP_040854203 | 100% | 241 | 77% | 80% |
| Sus scrofa | Wild boar | Artiodactyla | 94 | XP_003125804 | 96% | 248 | 79% | 82% |
| Myotis brandtii | Brandt's bat | Chiroptera | 94 | XP_005880696 | 98% | 248 | 75% | 79% |
| Sorex araneus | Common shrew | Eulipotyphla | 94 | XP_004618165 | 76% | 261 | 70% | 73% |
| Orycteropus afer | Aardvark | Afrotheria | 102 | XP_007956474 | 98% | 270 | 73% | 76% |
| Echinops telfairi | Lesser hedgehog tenrec | Afrotheria | 102 | XP_004717741 | 91% | 262 | 73% | 75% |
| Dasypus novemcinctus | Nine-banded armadillo | Xenarthra | 102 | XP_004469783 | 100% | 257 | 71% | 76% |
| Trichosurus vulpecula | Common brushtail possum | Diprotodontia | 160 | XP_036609710 | 100% | 254 | 62% | 67% |
| Phascolarctos cinereus | Koala | Diprotodontia | 160 | XP_020847076 | 100% | 266 | 60% | 65% |
| Vombatus ursinus | Common wombat | Diprotodontia | 160 | XP_027726322 | 100% | 291 | 52% | 58% |
| Sarcophilus harrisii | Tasmanian devil | Dasyuromorphia | 160 | XP_003770670 | 99% | 268 | 52% | 60% |
| Dromiciops gliroides | Colocolo opossum | Microbiotheria | 160 | XP_043856143 | 100% | 317 | 51% | 57% |
| Monodelphis domestica | Gray short-tailed opossum | Didelphimorphia | 160 | XP_016285839 | 96% | 292 | 54% | 59% |
| Ornithorhynchus anatinus | Platypus | Monotremata | 180 | XP_028910439 | 98% | 244 | 50% | 56% |
| Tachyglossus aculeatus | Short-beaked echidna | Monotremata | 180 | XP_038624254 | 98% | 264 | 47% | 54% |
| Ranitomeya imitator | Mimic poison frog | Anura | 352 | CAF5025995 | 96% | 251 | 26% | 33% |
| Xenopus tropicalis | Western clawed frog | Anura | 352 | KAE8606393 | 54% | 296 | 23% | 33% |
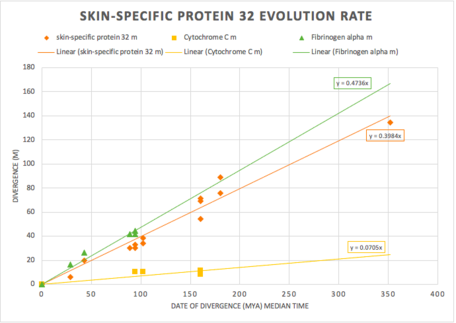
The figure below shows more information about the evolutionary rate of C1orf68 throughout its orthologs. The rate of evolution of C1orf68 was observed to be fast when comparing to cytochrome c and fibrinogen alpha. This observation is determined since C1orf68 appears to evolve at a similar rate to fibrinogen alpha, which serves as a standard for rapidly evolving genes.
Interacting proteins
Transcription factor binding sites
Three different transcription factors for C1orf68 were predicted and obtained from MatInspector Genomatics.[12]
GRHL2
Grainyhead-like 2 has been shown to impair keratinocyte differentiation through transcriptional inhibition of the gene in the epidermal differentiation complex.[21] Also showed enhanced protein and mRNA levels in chronic skin lesions, such as in psoriasis.[21]
ZEB1
Zinc finger E-box-binding homeobox 1 has been shown to regulate corneal epithelial terminal phenotype.[22]
GATA3
GATA-binding factor 3 has been shown localized in the cytoplasm and nucleus of proliferating keratinocytes but only in the nucleus in differentiated keratinocytes.[23] It has also been shown that GATA3 induces differentiation of primary keratinocytes, and suggested that it may regulate human interfollicular epidermal renewal.[23]
Protein-protein interactions
Other potential proteins that interact with C1orf68 are located in the table below. These proteins were selected from the results from prediction tools[4][24][25] because of their participation in the epidermal cornified envelope, the location of their gene within the epidermal differentiation complex, and the localization to the cytoplasm.
| Abbreviated Name | Full Name | Basis of Identification | Protein Description |
| KPRP | Keratinocyte proline-rich protein | Affinity Capture-MS[26] | This protein's gene is located on the epidermal differentiation complex on chromosome 1q21. Protein has a potential role in keratinocyte differentiation.[27] |
| TGM3 | Transglutaminase 3 | Affinity Capture-MS[26] | An epidermal cross-linking enzyme, it's involved with the formation of the cornfield envelope.[28] |
| CYLD | CYLD Lysine 63 Deubiquitinase | Affinity chromatography technology[29] | This protein functions as a deubiquitinating enzyme, and is localized in the cytoplasm.[30] |
Clinical significance
Psoriasis
C1orf68 is expressed differently when we look at samples of healthy skin, skin with psoriasis without lesions and skin with psoriasis with lesions.[31] In one study, it was suggested that proteins with significant differences in expression in skin with psoriasis without lesions and skin with psoriasis with lesions, could contribute to maintaining the non-lesional state and may add to our understanding of lesion formation.[32]
References
- ↑ Edqvist, Per-Henrik D.; Fagerberg, Linn; Hallström, Björn M.; Danielsson, Angelika; Edlund, Karolina; Uhlén, Mathias; Pontén, Fredrik (2014-11-19). "Expression of Human Skin-Specific Genes Defined by Transcriptomics and Antibody-Based Profiling". Journal of Histochemistry & Cytochemistry 63 (2): 129–141. doi:10.1369/0022155414562646. ISSN 0022-1554. PMID 25411189. PMC 4305515. http://dx.doi.org/10.1369/0022155414562646.
- ↑ Marshall, D.; Hardman, M. J.; Nield, K. M.; Byrne, C. (2001-11-06). "Differentially expressed late constituents of the epidermal cornified envelope". Proceedings of the National Academy of Sciences 98 (23): 13031–13036. doi:10.1073/pnas.231489198. ISSN 0027-8424. PMID 11698679. Bibcode: 2001PNAS...9813031M.
- ↑ Toulza, Eve; Mattiuzzo, Nicolas R; Galliano, Marie-Florence; Jonca, Nathalie; Dossat, Carole; Jacob, Daniel; de Daruvar, Antoine; Wincker, Patrick et al. (2007). "Large-scale identification of human genes implicated in epidermal barrier function". Genome Biology 8 (6): R107. doi:10.1186/gb-2007-8-6-r107. ISSN 1465-6906. PMID 17562024.
- ↑ 4.0 4.1 4.2 4.3 "Homo Sapiens C1orf68". https://www.ncbi.nlm.nih.gov/gene/100129271.
- ↑ "BLAT Results". https://genome.ucsc.edu/cgi-bin/hgBlat.
- ↑ "skin-specific protein 32 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_001019850.1.
- ↑ "Anti-C1ORF68 Rabbit Polyclonal Antibody" (in en). https://sg.vwr.com/store/product/16379657/anti-c1orf68-rabbit-polyclonal-antibody.
- ↑ 8.0 8.1 "ExPASy - Compute pI/Mw tool". https://web.expasy.org/compute_pi/.
- ↑ "SAPS < Sequence Statistics < EMBL-EBI". https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ "Motif Scan" (in en). https://myhits.sib.swiss/cgi-bin/motif_scan.
- ↑ "Dotlet JS". https://dotlet.vital-it.ch/.
- ↑ 12.0 12.1 12.2 "Genomatix: Login Page". http://www.genomatix.de/cgi-bin/sessions/login.pl?s=692b50cf896e42c54ccd6e36c5b751c9.
- ↑ "Tissue expression of C1orf68 - Summary - The Human Protein Atlas". https://www.proteinatlas.org/ENSG00000198854-C1orf68/tissue.
- ↑ "PAXdb: Protein Abundance Database". https://pax-db.org/search?q=c1orf68&speciesId=.
- ↑ "Single cell type - C1orf68 - The Human Protein Atlas". https://www.proteinatlas.org/ENSG00000198854-C1orf68/single+cell+type.
- ↑ "Anti-C1orf68 Antibody (HPA040836) - Atlas Antibodies". https://www.atlasantibodies.com/products/antibodies/primary-antibodies/triple-a-polyclonals/c1orf68-antibody-hpa040836/.
- ↑ 17.0 17.1 "RNA Folding Form". http://www.unafold.org/mfold/applications/rna-folding-form.php.
- ↑ "Subcellular - C1orf68 - The Human Protein Atlas". https://www.proteinatlas.org/ENSG00000198854-C1orf68/subcellular.
- ↑ Holthaus, K (Jun 2021). "Gene duplications and gene loss in the epidermal differentiation complex during the evolutionary land-to-water transition of Cetaceans". Scientific Reports 11 (1): 12334. doi:10.1038/s41598-021-91863-3. PMID 34112911.
- ↑ 20.0 20.1 "TimeTree :: The Timescale of Life". http://www.timetree.org/.
- ↑ 21.0 21.1 Chen, W; Xiao Liu, Z; Oh, J-E; Shin, K-H; Kim, R H; Jiang, M; Park, N-H; Kang, M K (2012). "Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism". Cell Death & Disease 3 (12): e450. doi:10.1038/cddis.2012.190. ISSN 2041-4889. PMID 23254293. PMC 3542624. http://dx.doi.org/10.1038/cddis.2012.190.
- ↑ Ortiz-Melo, María Teresa; Garcia-Murillo, Maria Jimena; Salazar-Rojas, Víctor Manuel; Campos, Jorge E.; Castro-Muñozledo, Federico (2021). "Transcriptional profiles along cell programming into corneal epithelial differentiation". Experimental Eye Research 202: 108302. doi:10.1016/j.exer.2020.108302. ISSN 0014-4835. PMID 33098888. http://dx.doi.org/10.1016/j.exer.2020.108302.
- ↑ 23.0 23.1 Masse, Ingrid; Barbollat-Boutrand, Laetitia; Kharbili, Manale El; Berthier-Vergnes, Odile; Aubert, Damien; Lamartine, Jérôme (2013-12-18). "GATA3 inhibits proliferation and induces expression of both early and late differentiation markers in keratinocytes of the human epidermis". Archives of Dermatological Research 306 (2): 201–208. doi:10.1007/s00403-013-1435-5. ISSN 0340-3696. PMID 24346062. http://dx.doi.org/10.1007/s00403-013-1435-5.
- ↑ "BioGRID | Database of Protein, Chemical, and Genetic Interactions". https://thebiogrid.org/.
- ↑ "GeneMANIA". https://genemania.org/.
- ↑ 26.0 26.1 Marcon, Edyta; Ni, Zuyao; Pu, Shuye; Turinsky, Andrei L; Trimble, Sandra Smiley; Olsen, Jonathan B; Silverman-Gavrila, Rosalind; Silverman-Gavrila, Lorelei et al. (2014). "Human Chromatin Related Protein Interactions Identify a Demethylase Complex Required for Chromosome Segregation". Cell Reports 8 (1): 297–310. doi:10.1016/j.celrep.2014.05.050. ISSN 2211-1247. PMID 24981860.
- ↑ Lee, Woong-Hee; Jang, Sunhyae; Lee, Jung-Suk; Lee, Young; Seo, Eun-Young; You, Kwan-Hee; Lee, Seung-Chul; Nam, Kwang-Il et al. (2005). "Molecular Cloning and Expression of Human Keratinocyte Proline-Rich Protein (hKPRP), an Epidermal Marker Isolated from Calcium-Induced Differentiating Keratinocytes". Journal of Investigative Dermatology 125 (5): 995–1000. doi:10.1111/j.0022-202x.2005.23887.x. ISSN 0022-202X. PMID 16297201.
- ↑ de Koning, H.D.; van den Bogaard, E.H.; Bergboer, J.G.M.; Kamsteeg, M.; van Vlijmen-Willems, I.M.J.J.; Hitomi, K.; Henry, J.; Simon, M. et al. (2012-05-25). "Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair". British Journal of Dermatology 166 (6): 1245–1254. doi:10.1111/j.1365-2133.2012.10885.x. ISSN 0007-0963. PMID 22329734. http://dx.doi.org/10.1111/j.1365-2133.2012.10885.x.
- ↑ Elliott, Paul R.; Leske, Derek; Hrdinka, Matous; Bagola, Katrin; Fiil, Berthe K.; McLaughlin, Stephen H.; Wagstaff, Jane; Volkmar, Norbert et al. (2016). "SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling". Molecular Cell 63 (6): 990–1005. doi:10.1016/j.molcel.2016.08.001. ISSN 1097-2765. PMID 27591049. PMC 5031558. http://dx.doi.org/10.1016/j.molcel.2016.08.001.
- ↑ "GeneCard-CYLD". https://www.genecards.org/cgi-bin/carddisp.pl?gene=CYLD.
- ↑ "Psoriasis lesional and non-lesional skin - - GEO DataSets - NCBI". https://www.ncbi.nlm.nih.gov/gds?LinkName=geoprofiles_gds&from_uid=100698577.
- ↑ Szél, Edit; Bozó, Renáta; Hunyadi-Gulyás, Éva; Manczinger, Máté; Szabó, Kornélia; Kemény, Lajos; Bata-Csörgő, Zsuzsanna; Groma, Gergely (2019-08-06). "Comprehensive Proteomic Analysis Reveals Intermediate Stage of Non-Lesional Psoriatic Skin and Points out the Importance of Proteins Outside this Trend". Scientific Reports 9 (1): 11382. doi:10.1038/s41598-019-47774-5. ISSN 2045-2322. PMID 31388062. PMC 6684579. Bibcode: 2019NatSR...911382S. http://dx.doi.org/10.1038/s41598-019-47774-5.
 |