Biology:Deubiquitinating enzyme
Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, or ubiquitin isopeptidases, are a large group of proteases[1] that cleave ubiquitin from proteins.[2] Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions.[3][4][5] DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.[2]
Classes
In humans there are 102 putative DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases, consisting of 58 ubiquitin-specific proteases (USPs), 4 ubiquitin C-terminal hydrolases (UCHs), 5 Machado-Josephin domain proteases (MJDs), 14 ovarian tumour proteases (OTU), and 14 Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain-containing genes. 11 of these proteins are predicted to be non-functional, leaving 79 functional enzymes.[6] In yeast, the USPs are known as ubiquitin-specific-processing proteases (UBPs).
Cysteine proteases
There are six main superfamilies of cysteine protease DUBs:[7]
- the ubiquitin-specific protease (USP/UBP) superfamily; (USP1, USP2, USP3, USP4, USP5, USP6, USP7, USP8, USP9X, USP9Y, USP10, USP11, USP12, USP13, USP14, USP15, USP16, USP17, USP17L2, USP17L3, USP17L4, USP17L5, USP17L7, USP17L8, USP18, USP19, USP20, USP21, USP22, USP23, USP24, USP25, USP26, USP27X, USP28, USP29, USP30, USP31, USP32, USP33, USP34, USP35, USP36, USP37, USP38, USP39, USP40, USP41, USP42, USP43, USP44, USP45, USP46)
- the ovarian tumour (OTU) superfamily (OTUB1, OTUB2);
- and the Machado-Josephin domain (MJD) superfamily. (ATXN3, ATXN3L)
- the ubiquitin C-terminal hydrolase (UCH) superfamily; (BAP1, UCHL1, UCHL3, UCHL5)
- the MINDY family of K48-specific deubiquitinases; (MINDY1, MINDY2, MINDY3, MINDY4)[8]
- the recently discovered ZUFSP family, at present solely represented by ZUP1[9][10]
| UCH | |||||||||
|---|---|---|---|---|---|---|---|---|---|
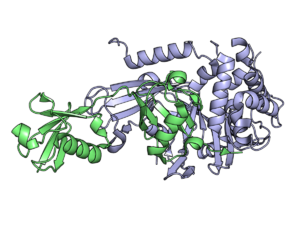
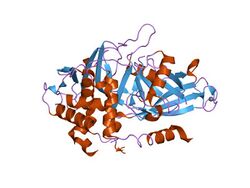 USP2 in complex with ubiquitin. | |||||||||
| Identifiers | |||||||||
| Symbol | UCH | ||||||||
| Pfam | PF00443 | ||||||||
| Pfam clan | CL0125 | ||||||||
| InterPro | IPR001394 | ||||||||
| PROSITE | PDOC00750 | ||||||||
| MEROPS | C19 | ||||||||
| SCOP2 | 1nb8 / SCOPe / SUPFAM | ||||||||
| |||||||||
There is also a little known putative group of DUBs called the permutated papain fold peptidases of dsDNA viruses and eukaryote (PPPDEs) superfamily, which, if shown to be bona fide DUBs, would be the seventh in the cysteine protease class.[11]
Metalloproteases
The Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain superfamily proteins bind zinc and hence are metalloproteases.[7]
Role of deubiquitinating enzymes
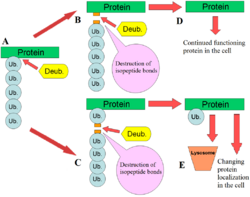
DUBs play several roles in the ubiquitin pathway. One of the best characterised functions of DUBs is the removal of monoubiqutin and polyubiquitin chains from proteins. These modifications are a post translational modification (addition to a protein after it has been made) where single ubiquitin proteins or chains of ubiquitin are added to lysines of a substrate protein. These ubiquitin modifications are added to proteins by the ubiquitination machinery; ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). The end result is ubiquitin bound to lysine residues via an isopeptide bond.[12] Proteins are affected by these modifications in a number of ways: they regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions.[3][4][5] DUBs play the antagonistic role in this axis by removing these modifications, therefore reversing the fate of the proteins.[2] In addition, a less understood role of DUBs is the cleavage of ubiquitin-like proteins such as SUMO and NEDD8. Some DUBs may have the ability to cleave isopeptide bonds between these proteins and substrate proteins.[13]
They activate ubiquitin by the proteolysis (breaking down) of the inactive expressed forms of ubiquitin. Ubiquitin is encoded in mammals by 4 different genes: UBA52, RPS27A, UBB and UBC. A similar set of genes is found in other eukaryotes such as yeast. The UBA52 and RPS27A genes produce ubiquitin that is fused to ribosomal proteins and the UBB and UBC genes produce polyubiquitin (a chain of ubiquitin joined by their C- and N-termini).[14][15] DUBs cleave the ubiquitin from these proteins, producing active single units of ubiquitin.[2]
DUBs also cleave single ubiquitin proteins that may have had their C-terminal tails accidentally bound to small cellular nucleophiles.[2] These ubiquitin-amides and ubiquitin-thioesters may be formed during standard ubiquitination reactions by the E1-E2-E3 cascade. Glutathione and polyamines are two nucleophiles that might attack the thiolester bond between ubiquitin and these enzymes. Ubiquitin C-terminal hydrolase is an example of the DUB that hydrolyses these bonds with broad specificity.[13][16]
Free polyubiquitin chains are cleaved by DUBs to produce monoubiquitin. The chains may be produced by the E1-E2-E3 machinery in the cell free from any substrate protein. Another source of free polyubiquitin is the product of ubiquitin-substrate cleavage. If DUBs cleave the base of the polyubiquitin chain that is attached to a protein, the whole chain will become free and needs to be recycled by DUBs.[2]
Domains
DUBs often contain a catalytic domain surrounded by one or more accessory domains, some of which contribute to target recognition. These additional domains include domain present in ubiquitin-specific proteases (DUSP) domain; ubiquitin-like (UBL) domain; meprin and TRAF homology (MATH) domain; zinc-finger ubiquitin-specific protease (ZnF-UBP) domain; zinc-finger myeloid, nervy and DEAF1 (ZnF-MYND) domain; ubiquitin-associated (UBA) domain; CHORD-SGT1 (CS) domain; microtubule-interacting and trafficking (MIT) domain; rhodenase-like domain; TBC/RABGAP domain; and B-box domain.[6][17]
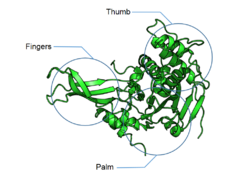
Catalytic domain
The catalytic domain of DUBs is what classifies them into particular groups; USPs, OTUs, MJDs, UCHs and MPN+/JAMMs. The first 4 groups are cysteine proteases, whereas the latter is a zinc metalloprotease. The cysteine protease DUBs are papain-like and thus have a similar mechanism of action. They use either catalytic dyads or triads (either two or three amino acids) to catalyse the hydrolysis of the amide bonds between ubiquitin and the substrate. The active site residues that contribute to the catalytic activity of the cysteine protease DUBs are cysteine (dyad/triad), histidine (dyad/triad) and aspartate or asparagine (triad only). The histidine is polarised by the aspartate or asparagine in catalytic triads or by other ways in dyads. This polarised residue lowers the pKa of the cysteine, allowing it to perform a nucleophilic attack on the isopeptide bond between the ubiquitin C-terminus and the substrate lysine. Metalloproteases coordinate zinc ions with histidine, aspartate and serine residues, which activate water molecules and allows them to attack the isopeptide bond.[18][19]
UBL
Ubiquitin-like (UBL) domains have a similar structure (fold) to ubiquitin, except they lack the terminal glycine residues. 18 USPs are proposed to have UBL domains. Only 2 other DUBs have UBLs outside the USP group: OTU1 and VCPIP1. USP4, USP7, USP11, USP15, USP32, USP40 and USP47 have multiple UBL domains. Sometimes the UBL domains are in tandem, such as in USP7 where 5 tandem C-terminal UBL domains are present. USP4, USP6, USP11, USP15, USP19, USP31, USP32 and USP43 have UBL domains inserted into the catalytic domain. The functions of UBL domains are different between USPs, but commonly they regulate USP catalytic activity. They can coordinate localisation at the proteasome (USP14); negatively regulate USPs by competing for the catalytic site of the USP (USP4), and induce conformational changes to increase catalytic activity (USP7).[17][20][21] Like other UBL domains, the structure of USP UBL domains show a β-grasp fold.[22][23]
DUSP
| DUSP domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
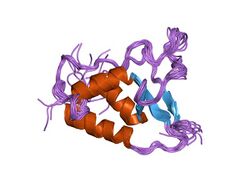 Solution structure of the DUSP domain of HUSP15. | |||||||||
| Identifiers | |||||||||
| Symbol | DUSP | ||||||||
| Pfam | PF06337 | ||||||||
| InterPro | IPR006615 | ||||||||
| MEROPS | C19 | ||||||||
| |||||||||
Single or multiple tandem DUSP domains of approximately 120 residues are found in six USPs. The function of the DUSP domain is currently unknown but it may play a role in protein-protein interaction, in particular to DUBs substrate recognition. This is predicted because of the hydrophobic cleft present in the DUSP domain of USP15 and that some protein interactions with DUSP containing USPs do not occur without these domains. The DUSP domain displays a novel tripod-like fold comprising three helices and an anti-parallel beta-sheet made of three strands. This fold resembles the legs (helices) and seat (beta-sheet) of the tripod. Within most DUSP domains in USPs there is a conserved sequence of amino acids known as the PGPI motif. This is a sequence of four amino acids; proline, glycine, proline and isoleucine, which packs against the three-helix bundle and is highly ordered.[6][24]
Role in disease
The full extent of the role of DUBs in diseases remains to be elucidated. Their involvement in disease is predicted due to known roles in physiological processes that are involved in disease states; including cancer and neurological disorders.[25]
The enzyme USP28 is over-expressed in different types of cancer such as colon or lung. In addition, USP28 deubiquitinates and stabilizes important oncogenes such as c-Myc, Notch1, c-jun or ΔNp63.[26][27][28] In squamous tumors, USP28 regulates the resistance to chemotherapy regulating DNA repair via ΔNp63-Fanconia anemia pathway axis.[29]
The deubiquitinating enzymes UCH-L3 and YUH1 are able to hydrolyse mutant ubiquitin UBB+1 despite the fact that the glycine at position 76 is mutated.[30]
UCH-L1 levels are high in various types of malignancies (cancer).[31]
Role in the cell cycle
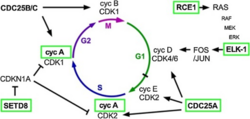
DUBs play an active role in modulating the cell cycle. Ubiquitin-specific-processing protease (USP) is a family of deubiquitinating enzymes that play a crucial role in cell cycle regulation.[32] Two such enzymes include USP17 and USP44. USP17 regulates pathways responsible for progressing cells through the cell cycle.[33] Its targets include regulators of Ras, CDK2, and Cyclin A.[34] USP44 plays an important role in anaphase initiation.[35] New research into the mitotic checkpoint has revealed a novel role for USP44 in regulating cell cycle progression.[35]
USP regulation of Ras
The ERK Pathway allows for the transduction of external mitogenic signals into intracellular signals promoting cellular proliferation. One of the key regulators of this pathways is Ras, a GTPase that, upon activation, binds GTP to “turn on” the subsequent signaling cascade. Ras converting enzyme 1 (RCE1) post-translationally cleaves the 3 residues on the C-terminus of Ras, allowing Ras to properly localize to the plasma membrane.[36]
USP17 acts to deubiquitinate K63-ubiquitin domains on RCE1.[34] Such stabilization of RCE1 allows for proper localization of Ras, thus promoting proliferation upon activation of early receptors in the ERK Pathway. Ras hyperactivity can result in cell cycle dysregulation.[37] Thus, regulation of Ras through USP17 acts as another point in Ras regulation.
USP regulation of G1-S transition
Cyclin-dependent kinases (CDKs) are a family of enzymes that phosphorylate serine and threonine residues to drive the cell through the cell cycle. Activation of CDK2 is critical for the G1-S transition. For CDK2 to be activated, cyclin A must bind to the cyclin-dependent kinase complex (CDKC). Cell division cycle 25A (CDC25A) is a phosphatase that removes an inhibitory phosphate group from CDK2.[38] While ubiquitination would mark CDC25A for degradation, thus blocking progression to S phase, USP17 deubiquitinates CDC25A.[34] An increase in CDC25A stability promotes CDKC activity, thus driving the cell through the G1-S transition.
USP17 also regulates cell cycle progression by acting on SETD8 to downregulate transcription of cyclin-dependent kinase inhibitor 1 (CDKN1A), also known as p21.[34] CDKN1A binds to and inhibits CDK2 using its N-terminal binding domain, thus blocking progression through the G1-S transition. SETD8, a methyltransferase, uses S-Adenosyl methionine to methylate the Lys20 residue of histone 4, resulting in the condensation of chromosomes.[39] This compaction of the DNA downregulates CDKN1A transcription. USP17 deubiquitinates SETD8, thus reducing its propensity for degradation and increasing its intracellular stability.[34] The resulting downregulation in CDKN1A transcription promotes CDK2 activity, allowing the cell to progress through the G1-S transition. See schematic of the role of DUBs in the cell cycle regulation.[34]
USP44 in anaphase initiation
The spindle checkpoint (also referred to as the mitotic checkpoint) ensures proper separation of chromosomes. Broadly, the mitotic checkpoint promotes fidelity in chromosomal segregation, increasing the likelihood that each daughter cell receives only one duplicated chromosome.[40] Such a mechanism is crucial, as errors in chromosomal separation have been implicated in cancer, birth defects, and antibiotic resistance in pathogens.[41] One of the core regulator proteins is the anaphase-promoting complex (APC/C). APC/C ubiquitinates securin.[42] The resulting destruction of securing release separase,[40] which hydrolyzes cohesion – the protein that binds sister chromatids together.
New research from Stegmeier and colleagues[35] published in the journal Nature demonstrates a crucial role for USP44 in regulating the spindle checkpoint. Using an shRNA screen, USP44 was identified to stabilize the inhibition of APC/C[35] The binding of CDC20 to APC/C is required for the ubiquitination of securin.[43] A protein called hMAD2 can form an inactive trimer with APC and CDC20, forming the hMAD2-CDC-APC complex.[43] Upon the ubiquitination of CDC20 by UbcH10, hMAD2 dissociates, and APC/C becomes active.[44] It is important to note that ubiquitination of CDC20 does not serve to mark it for degradation, but rather promote dissociation of hMAD2 from the hMAD2-CDC-APC complex. USP44, a ubiquitin-specific-processing protease, can stabilize the inactive hMAD2-CDC-APC complex by counteracting UbcH10 ubiquitination. This blocks hMAD2 dissociation and allows for proper regulation of APC/C, keeping it inactive until proper attachment of the mitotic spindle. Upon proper attachment, switch-like behavior allows for the activation of APC/C.[35] This results in the cleavage of cohesion, allowing for the separation of sister chromatids.
Role in p53-mediated DNA damage repair
DNA damage can prove catastrophic for an organism. Mechanisms for DNA mutation include oxidative stress, DNA replication errors, exogenous carcinogens, radiation, and spontaneous base mutation. Upon DNA damage, cell cycle progression is halted to prevent propagation of the mutation. The TP53 gene (also known as p53) is crucial in ensuring the conservation of the genome.[45] Deubiquitinating enzymes play an integral role in maintaining p53’s function.
In healthy cells, p53 activates the E3 ubiquitin ligase MDM2 which in turn ubiquitinates p53. This creates a negative feedback loop, whereby the degradation of p53 allows for cells to flow through the cell cycle.[46][circular reference] Upon DNA damage, Ubiquitin-specific-processing protease 7 (USP7) stabilizes p53 by cleaving ubiquitin.[47] For USP7 to deubiquitinate p53, it must localize to the nucleus. However, no nuclear localization sequence (NLS) has been found.[48] Despite no known NLS, one study showed that, upon deletion of USP7’s N-terminus, no nuclear localization occurred.[48] It is possible that other proteins facilitate nuclear entry of USP7.
Once stabilized, p53 can exert its tumor suppression function. Downstream pathways of p53 act to either halt cell cycle progression in G1 or G2 phases of the cell cycle[49] or promote cell-death, depending on the severity of the DNA damage.[50] See schematic of the role of USP7 in the p53-dependent pathway.[49] or promote cell-death, depending on the severity of the DNA damage.[50] See schematic of the role of USP7 in the p53-dependent pathway.[50]
References
- ↑ "Regulation of ubiquitin-dependent processes by deubiquitinating enzymes". FASEB Journal 11 (14): 1245–56. December 1997. doi:10.1096/fasebj.11.14.9409543. PMID 9409543.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 "Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes". Annual Review of Biochemistry 78: 363–97. 2009. doi:10.1146/annurev.biochem.78.082307.091526. PMID 19489724.
- ↑ 3.0 3.1 "The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction". Physiological Reviews 82 (2): 373–428. April 2002. doi:10.1152/physrev.00027.2001. PMID 11917093.
- ↑ 4.0 4.1 "Proteasome-independent functions of ubiquitin in endocytosis and signaling". Science 315 (5809): 201–5. January 2007. doi:10.1126/science.1127085. PMID 17218518. Bibcode: 2007Sci...315..201M.
- ↑ 5.0 5.1 "Non-traditional functions of ubiquitin and ubiquitin-binding proteins". The Journal of Biological Chemistry 278 (38): 35857–60. September 2003. doi:10.1074/jbc.R300018200. PMID 12860974.
- ↑ 6.0 6.1 6.2 "A genomic and functional inventory of deubiquitinating enzymes". Cell 123 (5): 773–86. December 2005. doi:10.1016/j.cell.2005.11.007. PMID 16325574.
- ↑ 7.0 7.1 "Mechanism and function of deubiquitinating enzymes". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1695 (1–3): 189–207. November 2004. doi:10.1016/j.bbamcr.2004.10.003. PMID 15571815.
- ↑ "MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes". Molecular Cell 63 (1): 146–55. July 2016. doi:10.1016/j.molcel.2016.05.009. PMID 27292798.
- ↑ "Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability". Molecular Cell 70 (1): 150–164.e6. April 2018. doi:10.1016/j.molcel.2018.02.023. PMID 29576527.
- ↑ Hermanns, Thomas; Pichlo, Christian; Woiwode, Ilka; Klopffleisch, Karsten; Witting, Katharina F.; Ovaa, Huib; Baumann, Ulrich; Hofmann, Kay (2018-02-23). "A family of unconventional deubiquitinases with modular chain specificity determinants" (in en). Nature Communications 9 (1): 799. doi:10.1038/s41467-018-03148-5. ISSN 2041-1723. PMID 29476094.
- ↑ "Novel predicted peptidases with a potential role in the ubiquitin signaling pathway". Cell Cycle 3 (11): 1440–50. November 2004. doi:10.4161/cc.3.11.1206. PMID 15483401.
- ↑ "Modification of proteins by ubiquitin and ubiquitin-like proteins". Annual Review of Cell and Developmental Biology 22: 159–80. 2006. doi:10.1146/annurev.cellbio.22.010605.093503. PMID 16753028.
- ↑ 13.0 13.1 "Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway". The International Journal of Biochemistry & Cell Biology 35 (5): 590–605. May 2003. doi:10.1016/s1357-2725(02)00392-8. PMID 12672452.
- ↑ "Regulatory mechanisms involved in the control of ubiquitin homeostasis". Journal of Biochemistry 147 (6): 793–8. June 2010. doi:10.1093/jb/mvq044. PMID 20418328.
- ↑ "The yeast ubiquitin genes: a family of natural gene fusions". The EMBO Journal 6 (5): 1429–39. May 1987. doi:10.1002/j.1460-2075.1987.tb02384.x. PMID 3038523.
- ↑ "Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides". The Journal of Biological Chemistry 260 (13): 7903–10. July 1985. doi:10.1016/S0021-9258(17)39538-8. PMID 2989266.
- ↑ 17.0 17.1 "Breaking the chains: structure and function of the deubiquitinases". Nature Reviews. Molecular Cell Biology 10 (8): 550–63. August 2009. doi:10.1038/nrm2731. PMID 19626045.
- ↑ Komander D (2010). "Mechanism, Specificity and Structure of the Deubiquitinases". Conjugation and Deconjugation of Ubiquitin Family Modifiers. Subcellular Biochemistry. 54. 69–87. doi:10.1007/978-1-4419-6676-6_6. ISBN 978-1-4419-6675-9.
- ↑ "Emerging roles for cysteine proteases in human biology". Annual Review of Physiology 59: 63–88. 1997. doi:10.1146/annurev.physiol.59.1.63. PMID 9074757.
- ↑ "The role of UBL domains in ubiquitin-specific proteases". Biochemical Society Transactions 40 (3): 539–45. June 2012. doi:10.1042/BST20120004. PMID 22616864.
- ↑ "Dissection of USP catalytic domains reveals five common insertion points". Molecular BioSystems 5 (12): 1797–808. December 2009. doi:10.1039/b907669g. PMID 19734957.
- ↑ "Structural variability of the ubiquitin specific protease DUSP-UBL double domains". FEBS Letters 585 (21): 3385–90. November 2011. doi:10.1016/j.febslet.2011.09.040. PMID 22001210.
- ↑ "Structure of the USP15 N-terminal domains: a β-hairpin mediates close association between the DUSP and UBL domains". Biochemistry 50 (37): 7995–8004. September 2011. doi:10.1021/bi200726e. PMID 21848306.
- ↑ "Solution structure of the human ubiquitin-specific protease 15 DUSP domain". The Journal of Biological Chemistry 281 (8): 5026–31. February 2006. doi:10.1074/jbc.M510993200. PMID 16298993.
- ↑ "Deubiquitylating enzymes and disease". BMC Biochemistry 9 (Suppl 1): S3. October 2008. doi:10.1186/1471-2091-9-S1-S3. PMID 19007433.
- ↑ "The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer". The Journal of Clinical Investigation 124 (8): 3407–18. August 2014. doi:10.1172/JCI73733. PMID 24960159.
- ↑ Prieto-Garcia C, Hartmann O, Reissland M, Braun F, Fischer T, Walz S, et al. (Jun 2019). "The USP28-∆Np63 axis is a vulnerability of squamous tumours". bioRxiv 10.1101/683508.
- ↑ "Maintaining protein stability of ∆Np63 via USP28 is required by squamous cancer cells". EMBO Molecular Medicine 12 (4): e11101. April 2020. doi:10.15252/emmm.201911101. PMID 32128997.
- ↑ Prieto-Garcia C, Hartmann O, Reissland M, Fischer T, Maier CR, Rosenfeldt M, et al. (Sep 2020). "Inhibition of USP28 overcomes Cisplatin-Resistance of Squamous Tumors by Suppression of the Fanconi Anemia Pathway". bioRxiv 10.1101/2020.09.10.291278.
- ↑ "Mutant ubiquitin (UBB+1) associated with neurodegenerative disorders is hydrolyzed by ubiquitin C-terminal hydrolase L3 (UCH-L3)". FEBS Letters 585 (16): 2568–74. August 2011. doi:10.1016/j.febslet.2011.06.037. PMID 21762696.
- ↑ "The potential role of ubiquitin c-terminal hydrolases in oncogenesis". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 1806 (1): 1–6. August 2010. doi:10.1016/j.bbcan.2010.03.001. PMID 20302916.
- ↑ "USP7 Is a Master Regulator of Genome Stability". Frontiers in Cell and Developmental Biology 8 (717): 717. August 2020. doi:10.3389/fcell.2020.00717. PMID 32850836.
- ↑ "The ubiquitin-specific protease USP17 prevents cellular senescence by stabilizing the methyltransferase SET8 and transcriptionally repressing p21". The Journal of Biological Chemistry 294 (44): 16429–16439. November 2019. doi:10.1074/jbc.RA119.009006. PMID 31533987.
- ↑ 34.0 34.1 34.2 34.3 34.4 34.5 "USP17-mediated de-ubiquitination and cancer: Clients cluster around the cell cycle". The International Journal of Biochemistry & Cell Biology 130 (130): 105886. January 2021. doi:10.1016/j.biocel.2020.105886. ISSN 1357-2725. PMID 33227393. https://nottingham-repository.worktribe.com/preview/5129584/ACCEPTED%20-%20USP17-mediated%20de-ubiquitination%20and%20Cancer.pdf.
- ↑ 35.0 35.1 35.2 35.3 35.4 "Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities". Nature 446 (7138): 876–81. April 2007. doi:10.1038/nature05694. PMID 17443180. Bibcode: 2007Natur.446..876S.
- ↑ "Knockout of the RAS endoprotease RCE1 accelerates myeloid leukemia by downregulating GADD45b". Leukemia 35 (2): 606–609. February 2021. doi:10.1038/s41375-020-0859-0. PMID 32398789.
- ↑ "Targeting Ras in myeloid leukemias". Clinical Cancer Research 14 (8): 2249–52. April 2008. doi:10.1158/1078-0432.CCR-07-1005. PMID 18413813.
- ↑ "The role of Cdc25A in the regulation of cell proliferation and apoptosis". Anti-Cancer Agents in Medicinal Chemistry 12 (6): 631–9. July 2012. doi:10.2174/187152012800617678. PMID 22263797.
- ↑ "Cell cycle: Disposing of SETD8". Nature Reviews. Molecular Cell Biology 11 (12): 819. December 2010. doi:10.1038/nrm3020. PMID 21102605.
- ↑ 40.0 40.1 "The mitotic checkpoint complex (MCC): looking back and forth after 15 years". AIMS Molecular Science 3 (4): 597–634. October 2016. doi:10.3934/molsci.2016.4.597. PMID 28920074.
- ↑ "The Consequences of Chromosome Segregation Errors in Mitosis and Meiosis". Biology 6 (1): 12. February 2017. doi:10.3390/biology6010012. PMID 28208750.
- ↑ "Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation". Genes & Development 18 (1): 88–98. January 2004. doi:10.1101/gad.285404. PMID 14724179.
- ↑ 43.0 43.1 "The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation". Genes & Development 12 (12): 1871–83. June 1998. doi:10.1101/gad.12.12.1871. PMID 9637688.
- ↑ "Cell biology: the checkpoint brake relieved". Nature 446 (7138): 868–9. April 2007. doi:10.1038/446868a. PMID 17443175. Bibcode: 2007Natur.446..868P.
- ↑ "TP53 gene". U.S. National Library of Medicine. https://medlineplus.gov/genetics/gene/tp53/.
- ↑ "p53". https://en.wikipedia.org/wiki/P53.
- ↑ "Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization". Nature 416 (6881): 648–53. April 2002. doi:10.1038/nature737. PMID 11923872. Bibcode: 2002Natur.416..648L.
- ↑ 48.0 48.1 "Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization". The FEBS Journal 274 (16): 4256–70. August 2007. doi:10.1111/j.1742-4658.2007.05952.x. PMID 17651432.
- ↑ 49.0 49.1 "Phosphatases reverse p53-mediated cell cycle checkpoints". Proceedings of the National Academy of Sciences of the United States of America 111 (20): 7172–3. May 2014. doi:10.1073/pnas.1405663111. PMID 24808140. Bibcode: 2014PNAS..111.7172D.
- ↑ 50.0 50.1 50.2 "Regulating tumor suppressor genes: post-translational modifications". Signal Transduction and Targeted Therapy 5 (1): 90. June 2020. doi:10.1038/s41392-020-0196-9. PMID 32532965.
 |