Biology:Frequency (gene)
| Frequency clock protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | FRQ | ||||||||
| Pfam | PF09421 | ||||||||
| InterPro | IPR018554 | ||||||||
| |||||||||
The frequency (frq) gene encodes the protein frequency (FRQ) that functions in the Neurospora crassa circadian clock. The FRQ protein plays a key role in circadian oscillator, serving to nucleate the negative element complex in the auto regulatory transcription-translation negative feedback-loop (TTFL) that is responsible for circadian rhythms in N. crassa.[1] Similar rhythms are found in mammals, Drosophila and cyanobacteria. Recently, FRQ homologs have been identified in several other species of fungi.[2] Expression of frq is controlled by the two transcription factors white collar-1 (WC-1) and white collar-2 (WC-2) that act together as the White Collar Complex (WCC) and serve as the positive element in the TTFL. Expression of frq can also be induced through light exposure in a WCC dependent manner. Forward genetics has generated many alleles of frq resulting in strains whose circadian clocks vary in period length.
Discovery
The frq locus was discovered by Jerry F. Feldman. Feldman had been a graduate student with Colin Pittendrigh at Princeton and went to Caltech in 1967 to begin genetic screens for circadian clock mutants. The screening was aided by recent work that improved the expression of the rhythm in Neurospora. Colin Pittendrigh and his colleagues had confirmed in 1959 that the daily cycle of asexual development, described in Neurospora crassa earlier by Brandt,[3] was in fact due to regulation by a circadian clock.[4] In work published not long before Feldman arrived at Caltech, Malcolm L. Sargent, Winslow R. Briggs and Dow O. Woodward at Stanford University reported that overt expression of the developmental rhythm in conidiation was enhanced in a strain of Neurospora called Timex.[5] (This strain contained a mutation in the locus band (bd), later shown to encode a mildly hyperactive allele of ras-1 so strains are now known as ras-1[bd].[6] Because rhythms in strains that include ras-1[bd] are easier to detect, ras-1[bd] is often incorporated into strains used for studies of circadian biology in Neurospora.[6]). Outputs of the Neurospora circadian clock include carotenoid synthesis as well as the asexual spore formation seen on race tubes, and recent evidence suggests that thousands of genes are under circadian control.[7][2]
Feldman used nitrosoguanidine as a mutagen and used race tubes to screen individual strains surviving the mutagenesis for their circadian period length. Race tubes are long hollow glass tubes bent at either end to hold an agar growth medium. When Neurospora is inoculated at one end of a tube it will grow to the other end, and in constant darkness the daily circadian cycle of growth and development is manifest.[8] Although Feldman's screens were successful he was slow to publish so the identity of mutant genes frq[1], frq[2], and frq[3] were not reported until 1973.[9] In 1986, frq was cloned by Jay Dunlap and his colleagues using a strategy that involved a long chromosome walk and successful application of the then-untried strategy of rescuing an arrhythmic behavioral mutant through transformation of exogenous DNA arising from the chromosome walk. The success of this strategy and of the cloning of a clock gene sparked interest in further research and understanding of the N. crassa circadian clock.[10] The expression of frq was later shown to rhythmically cycle; furthermore, when strains of Neurospora were engineered in which frq expression could be driven from a region distinct from the resident wild type gene, it was found that FRQ repressed its own expression and that no level of constant expression could support a circadian clock.[11] These experiments were the first to manipulate the expression of a clock gene through means that did not themselves affect the clock and established that autoregulatory negative feedback giving rise to cyclical clock gene expression lay at the core of the circadian oscillator.
Structure and function
Reflecting its role as a core clock protein, deletion of the frq gene results in arrhythmicity, and in Neurospora, the only function of FRQ is in the circadian clock. The frq gene can be activated from two distinct cis-acting sequences in its promoter, a distal site, the clock-box, used in the context of circadian regulation, and a site close to the principal transcription start site that is used for light-induced expression (the proximal light-regulatory element or PLRE). These frq transcripts both have capacity to encode two FRQ proteins, a long form of 989 amino acids (lFRQ) and a short form of 890 amino acids (sFRQ); both lFRQ and sFRQ are required for strong rhythmicity although the clock is able to persist at certain temperatures, albeit with a weaker rhythmicity, with just one of the proteins present.[13] The choice of which protein is made is the result of temperature-dependent splicing of the primary transcript such that it includes or excludes the ATG start codon for lFRQ.[14] The two forms of FRQ provide the Neurospora clock a greater range of temperatures over which it can operate optimally. An increase in temperature leads to increased expression of lFRQ, while sFRQ is unaffected. Warmer temperatures induce more efficient splicing of an intron in the translation start site.[7] Because sFRQ favors a longer period than lFRQ, free running rhythms in wild type Neurospora are somewhat decreased with increased temperature.[7]
FRQ has also been shown to interact with several other proteins. It interacts at all times with FRH (FRQ-interacting RNA helicase; an essential DEAD box-containing RNA helicase in Neurospora) to form a FRQ/FRH complex (FFC).[15][16] FRQ also stably interacts with casein kinase 1 (CK1) although the strength of the interaction changes with time of day. Additional interactions with other kinases including PRD-4 (CHK2)[17] and casein kinase 2 (CKII) are known.
Structural prediction programs suggest that only a few regions of FRQ are likely to fold into stable structures, and consistent with this a variety of experimental data indicate that FRQ is an intrinsically disordered protein.[18] In the absence of its partner FRH, FRQ is very unstable. The myriad time-of-day specific phosphorylation that characterize FRQ are predicted to provide structure to this otherwise disordered protein. There is no known domain structure to FRQ because of its highly disordered structure.
Typically, proteins show a codon usage bias where they are more likely to choose synonymous codons that are more available in their tRNA pool. Neurospora crassa has a relatively strong codon usage bias compared to S. cerevisiae, a commonly used organism for codon-optimization analysis. However, because FRQ is an intrinsically disordered protein, it does not have demonstrate codon usage bias. In fact, when its codons are optimized, the protein loses its function and the clock is disturbed. This is not the case for cyanobacterial clock genes, kaiB and kaiC, which both led to more robust clock function.[19]
Regulation
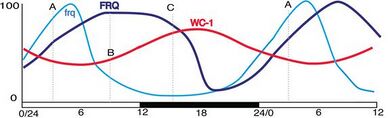
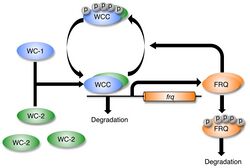
A description of the regulation of frq and FRQ requires a description of the clock cycle. The molecular basis of the circadian oscillator in Neurospora begins with two protein complexes. One is the FFC, the negative element complex composed of two copies of FRQ, FRH, and Casein kinase 1 as well as, probably, other less strongly bound proteins.[16] The other complex which acts as the positive element in the feedback loop includes WC-1 and WC-2; they are GATA transcription factors that, together, form the heterodimeric WCC via their PAS domains.[21] When WCC is released from the FFC negative element complex during subjective night, it binds to the clock-box within frequency (frq) gene promoter and activates frq transcription.[22][23] It has recently been shown that the Histone H3 Lysine 36 Methyltransferase, SET-2, is responsible for methylation of the frq gene to establish a chromatin state that will allow for transcription of frq by the WCC.[24]
The frequency (FRQ) protein accumulates and is progressively phosphorylated by CKI, CKII, and a calcium/calmodulin-dependent kinase (CAMK-1), and additional kinases, reaching its peak around mid-subjective day.[25][26][27] Kinase inhibitors reduce degradation of FRQ by preventing phosphorylation.[28] FRQ is phosphorylated at more than 100 sites based on in vitro analyses using mass spectrometry of lFRQ peptides. These sites appear within the protein in a highly reproducible manner indicating that the timing of the phosphorylations is important. Moreover, mutation of the sites shows that they work in domains, with some phosphorylations serving to lengthen period and others to shorten period.[16]
FRQ recruits kinases such as casein kinase 1a (CK-1a) that phosphorylate WCC, although the function of these phosphorylations is unclear as hyperphosphorylated WCC remains active. Eventually, repression is relieved when FRQ becomes so highly phosphorylated that the FFC no longer interacts with the WCC. This process occurs with a periodicity of around 22 hours in constant conditions.[29] At a later time, and with kinetics that do not influence the circadian cycle, this hyperphosphorylated FRQ is degraded through the ubiquitin/proteasome pathway. Heavily phosphorylated FRQ undergoes a conformational change that is detected by the FWD-1 protein, which is part of the SCF type E3 ligase.[30]
FRQ forms a homodimer via its coiled-coil domain located near the N-terminus. This dimerization is required for FRQ to interact with the WCC and repress its own expression.[31] Deletion of the WCC leads to an inability to form the homodimer, which causes frq to no longer be negatively regulated by FRQ concentration.[31] This leads to arrhythmicity.[31]
A positive feedback loop between FRQ and WCC has been proposed but details are not yet known. It is believed that WCC is degraded when it is transcriptionally active, and that prevention of this caused by the FFC allows for an accumulation of WCC.[32] This proposed mechanism has been shown to possibly be more complex in that FRQ may regulate WC-1 and WC-2 independently.[33] Recently the transcription factor ADV-1 was identified as a necessary transducer of clock outputs, including circadian rhythmicity in genes critical to somatic cell fusion.[34]
The frq gene is strongly induced by short duration exposure to light. Because the core of the clock is based on rhythmic expression of frq, acute light-induction provides a straightforward way to reset the clock.[35] Mammalian clocks are reset by light by a nearly identical mechanism, with mPer1 transcripts being induced by short flashes of light outside of the subjective day. The mPer1 mechanism in the mammalian clock draws closer similarities to the mechanism in Neurospora than to the mechanism of its homolog in Drosophila, per.[36]
Mutations
Forward genetics has been used to create Neurospora clock mutants with varied periods of conidiation. Although nine alleles have been described as having come from forward genetics, sequence analysis subsequent to the cloning of frq showed that frq[2] ,frq[4], and frq[6] shared the same single base change, and likewise frq[7] and frq[8] had the same single base change, so the redundant alleles have been dropped.[37] The periods of various frq mutants that arose from forward screens are as follows when measured at 25 °C, although because frq[3] and frq[7] result in clocks with altered temperature compensation, periods will be different at other temperatures: File:Importance-of-MAP-Kinases-during-Protoperithecial-Morphogenesis-in-Neurospora-crassa-pone.0042565.s006.ogv
| Mutant | frq[1] | frq[2] | frq[3 | frq[7] | frq[9] |
| Period (hr) | 16.5 | 19.3 | 24.0 | 29.0 | Arrhythmic |
FRQ-less oscillator (FLO)
A number of identifiably distinct oscillators outside of the FRQ/WCC system have been discovered; however, none of these FRQ-less oscillations (FLOs) satisfy the characteristics to be classified as circadian oscillators.[38] The circadian FRQ-WCC Oscillator (FWO) has been shown, via luciferase reporting, to continue running even when a FLO (the CDO or choline deficiency oscillator that controls conidiation under conditions of choline limitation) controls conidiation.[38] In the frq[9] mutant Neurospora crassa, a non-temperature compensated rhythm of conidiospore development was still observed in constant darkness (DD).[39] The period for frq null mutants varied from 12 to 35 hours but could be stabilized by the addition of farnesol or geraniol. However, this mechanism is not well understood.[40] Although this FRQ-less rhythm lost certain clock characteristics such as temperature compensation, temperature pulses were sufficient to reset the clock.[41] Another FLO is the NRO or Nitrate Reductase Oscillator that appears under conditions of nitrate starvation and is thought to arise from feedback loops within the nitrate assimilation pathway; it has a period length of about 24 hours but is not temperature compensated.[42] In short, there is much evidence to support FRQ-less oscillators in Neurospora crassa. One way to rationalize this is to assume that many are "slaves" to the frequency/white collar oscillator; they do not possess all of the characteristics of a circadian clock on their own because this is supplied by the FWO.[40] However, rhythms in clock-controlled gene-16 (ccg-16) are coupled to the FWO but function autonomously, demonstrating that Neurospora crassa contains at least 2 potential pacemakers, but only one that can be reset by light and temperature while maintaining temperature compensation.[40][43] The FRQ-less oscillator has never been proven to affect the true circadian clock.[43] The mechanism and significance for FRQ-less oscillators (FLO) are still under research.
Evolution
The FRQ protein is conserved within the Sordariacea but diverges outside of this group.[2][44] Nonetheless bona fide FRQ-based circadian cocks have been found in organisms other than Neurospora both within the Sordariacea, for instance, in the salient fungal pathogen Botrytis,[45] and also as far afield as Pyronema[46] within the Pezizomycetes, an early-diverging lineage of filamentous ascomycetes. Frq was even found in non-Dikarya group of fungi. The finding of frq and conserved circadian clock mechanism inside non-Dikarya, Arbuscular Mycorrhizal Fungi expanded the evolutionary history of this gene in Fungal kingdom.[47] frq seems to diverge very quickly during its evolution. A part of the reason why the FRQ primary amino acid sequence diverges so quickly may be because it is an intrinsically disordered protein and as a result lacks the structural constraints that limit sequence changes.[48][18] Since codon optimization of the frq gene results in impaired circadian feedback loop function, frq displays non-optimal codon usage bias across its open reading frame in contrast to most other genes.[49] FRQ is an intrinsically disordered protein that is not well conserved, even across fungi.[50] Unlike FRQ, however, WC-1 is very well conserved. It is the founding member of the family of blue light photoreceptors used in the entire Kingdom of fungi. Moreover, it is similar in structure and function to BMAL1. Casein kinase 2 is conserved in the circadian oscillators of plants (Arabidopsis) and flies (Drosophila).[30] A similar form of CKI is necessary for the degradation of period (PER) proteins in Drosophila and mammals.[30] The Drosophila gene slimb is orthologous to FWD1 in Neurospora, both of which are crucial for clock protein degradation.[30] In general, the TTFLs found in fungi and animals share a similar regulatory architecture, with a single step negative feedback loop, PAS-PAS heterodimeric activators that are conserved, and negative element proteins that largely lack structure and are much less well conserved. A similar palette of kinases modifies the clock proteins in all cases.
See also
References
- ↑ "The circadian clock of Neurospora crassa". FEMS Microbiology Reviews 36 (1): 95–110. January 2012. doi:10.1111/j.1574-6976.2011.00288.x. PMID 21707668.
- ↑ 2.0 2.1 2.2 "Around the Fungal Clock: Recent Advances in the Molecular Study of Circadian Clocks in Neurospora and Other Fungi". Advances in Genetics 92: 107–84. 2015-01-01. doi:10.1016/bs.adgen.2015.09.003. PMID 26639917.
- ↑ "Zonation in a prolinelcss strain of Neurospora". Mycologia 45 (2): 194–209. 1953. doi:10.1080/00275514.1953.12024261.
- ↑ Pittendrigh, C. S.; Bruce, V. G.; Rosensweig, N. S.; Rubin, M. L. (July 18, 1959). "Growth Patterns in Neurospora: A Biological Clock in Neurospora". Nature 184 (4681): 169–170. doi:10.1038/184169a0. Bibcode: 1959Natur.184..169P.
- ↑ "Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa". Plant Physiology 41 (8): 1343–9. October 1966. doi:10.1104/pp.41.8.1343. PMID 5978549.
- ↑ 6.0 6.1 "The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output". Genes & Development 21 (12): 1494–505. June 2007. doi:10.1101/gad.1551707. PMID 17575051.
- ↑ 7.0 7.1 7.2 "Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms". FEBS Letters 581 (30): 5759–64. December 2007. doi:10.1016/j.febslet.2007.11.043. PMID 18037381.
- ↑ "The circadian conidiation rhythm in Neurospora crassa". Seminars in Cell & Developmental Biology 7 (6): 765–774. December 1996. doi:10.1006/scdb.1996.0094.
- ↑ "Isolation of circadian clock mutants of Neurospora crassa". Genetics 75 (4): 605–13. December 1973. doi:10.1093/genetics/75.4.605. PMID 4273217.
- ↑ "The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period". Nature 339 (6225): 558–62. June 1989. doi:10.1038/339558a0. PMID 2525233. Bibcode: 1989Natur.339..558M.
- ↑ "Negative feedback defining a circadian clock: autoregulation of the clock gene frequency". Science 263 (5153): 1578–84. March 1994. doi:10.1126/science.8128244. PMID 8128244. Bibcode: 1994Sci...263.1578A.
- ↑ "Comprehensive modelling of the Neurospora circadian clock and its temperature compensation". PLOS Computational Biology 8 (3): e1002437. 2012. doi:10.1371/journal.pcbi.1002437. PMID 22496627. Bibcode: 2012PLSCB...8E2437T.
- ↑ "Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock". Cell 89 (3): 477–86. May 1997. doi:10.1016/S0092-8674(00)80228-7. PMID 9150147.
- ↑ "Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency". Molecular Biology of the Cell 16 (12): 5563–71. 2005. doi:10.1091/mbc.E05-08-0756. PMID 16195340.
- ↑ "Regulation of the Neurospora circadian clock by an RNA helicase". Genes & Development 19 (2): 234–41. January 2005. doi:10.1101/gad.1266805. PMID 15625191.
- ↑ 16.0 16.1 16.2 "Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock". Molecular Cell 34 (3): 354–63. 2009. doi:10.1016/j.molcel.2009.04.023. PMID 19450533.
- ↑ "The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles". Science 313 (5787): 644–9. 2006. doi:10.1126/science.1121716. PMID 16809488. Bibcode: 2006Sci...313..644P.
- ↑ 18.0 18.1 "Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ". Molecular Cell 52 (6): 832–43. December 2013. doi:10.1016/j.molcel.2013.11.005. PMID 24316221.
- ↑ Zhou, Mian; Wang, Tao; Fu, Jingjing; Xiao, Guanghua; Liu, Yi (2017-04-27). "Non-optimal codon usage influences protein structure in intrinsically disordered regions". Molecular Microbiology 97 (5): 974–987. doi:10.1111/mmi.13079. ISSN 0950-382X. PMID 26032251.
- ↑ "A circadian clock in Neurospora: how genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day". Cold Spring Harbor Symposia on Quantitative Biology 72: 57–68. 2007. doi:10.1101/sqb.2007.72.072. PMID 18522516.
- ↑ "Role of a white collar-1-white collar-2 complex in blue-light signal transduction". The EMBO Journal 18 (18): 4961–8. September 1999. doi:10.1093/emboj/18.18.4961. PMID 10487748.
- ↑ "White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter". Science 297 (5582): 815–9. 2002. doi:10.1126/science.1073681. PMID 12098706. Bibcode: 2002Sci...297..815F.
- ↑ "WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora". The EMBO Journal 20 (1–2): 109–17. January 2001. doi:10.1093/emboj/20.1.109. PMID 11226161.
- ↑ "Suppression of WHITE COLLAR-independent frequency Transcription by Histone H3 Lysine 36 Methyltransferase SET-2 Is Necessary for Clock Function in Neurospora". The Journal of Biological Chemistry 291 (21): 11055–63. May 2016. doi:10.1074/jbc.M115.711333. PMID 27002152.
- ↑ "Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY". Cell 89 (3): 469–76. 1997. doi:10.1016/S0092-8674(00)80227-5. PMID 9150146.
- ↑ "The Neurospora crassa circadian clock". Advances in Genetics 58: 25–66. 2007. doi:10.1016/s0065-2660(06)58002-2. ISBN 9780123738820. PMID 17452245.
- ↑ "Mechanism of the Neurospora circadian clock, a FREQUENCY-centric view". Biochemistry 54 (2): 150–6. January 2015. doi:10.1021/bi5005624. PMID 25302868.
- ↑ "Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock". Proceedings of the National Academy of Sciences of the United States of America 97 (1): 234–9. January 2000. doi:10.1073/pnas.97.1.234. PMID 10618401. Bibcode: 2000PNAS...97..234L.
- ↑ "Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination". Science 347 (6221): 1257277. January 2015. doi:10.1126/science.1257277. PMID 25635104.
- ↑ 30.0 30.1 30.2 30.3 "FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation". The EMBO Journal 22 (17): 4421–30. September 2003. doi:10.1093/emboj/cdg425. PMID 12941694.
- ↑ 31.0 31.1 31.2 "Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora". The EMBO Journal 20 (1–2): 101–8. January 2001. doi:10.1093/emboj/20.1.101. PMID 11226160.
- ↑ "FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock". Genetics 184 (2): 351–61. 2010. doi:10.1534/genetics.109.111393. PMID 19948888.
- ↑ Brody, Stuart, ed (2011-01-01). "The genetics of circadian rhythms in Neurospora". Advances in Genetics 74: 55–103. doi:10.1016/b978-0-12-387690-4.00003-9. ISBN 9780123876904. PMID 21924975.
- ↑ Dekhang, Rigzin; Wu, Cheng; Smith, Kristina M.; Lamb, Teresa M.; Peterson, Matthew; Bredeweg, Erin L.; Ibarra, Oneida; Emerson, Jillian M. et al. (2017-01-05). "The Neurospora Transcription Factor ADV-1 Transduces Light Signals and Temporal Information to Control Rhythmic Expression of Genes Involved in Cell Fusion". G3: Genes, Genomes, Genetics 7 (1): 129–142. doi:10.1534/g3.116.034298. ISSN 2160-1836. PMID 27856696.
- ↑ "Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript". Cell 81 (7): 1003–12. 1995. doi:10.1016/S0092-8674(05)80005-4. PMID 7600569.
- ↑ "Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript". Cell 91 (7): 1043–53. December 1997. doi:10.1016/s0092-8674(00)80494-8. PMID 9428526.
- ↑ "Circadian clock locus frequency: protein encoded by a single open reading frame defines period length and temperature compensation". Proceedings of the National Academy of Sciences of the United States of America 91 (16): 7683–7. 1994. doi:10.1073/pnas.91.16.7683. PMID 8052643. Bibcode: 1994PNAS...91.7683A.
- ↑ 38.0 38.1 "A developmental cycle masks output from the circadian oscillator under conditions of choline deficiency in Neurospora". Proceedings of the National Academy of Sciences of the United States of America 104 (50): 20102–7. December 2007. doi:10.1073/pnas.0706631104. PMID 18056807. Bibcode: 2007PNAS..10420102S.
- ↑ "A recessive circadian clock mutation at the frq locus of Neurospora crassa". Genetics 114 (4): 1095–110. December 1986. doi:10.1093/genetics/114.4.1095. PMID 2948874.
- ↑ 40.0 40.1 40.2 "Circadian rhythms from multiple oscillators: lessons from diverse organisms". Nature Reviews Genetics 6 (7): 544–56. July 2005. doi:10.1038/nrg1633. PMID 15951747.
- ↑ "Circadian rhythms in Neurospora crassa: farnesol or geraniol allow expression of rhythmicity in the otherwise arrhythmic strains frq10, wc-1, and wc-2". Journal of Biological Rhythms 18 (4): 287–96. August 2003. doi:10.1177/0748730403255934. PMID 12932081.
- ↑ "A nitrate-induced frq-less oscillator in Neurospora crassa". Journal of Biological Rhythms 19 (4): 280–6. 2004. doi:10.1177/0748730404265532. PMID 15245647.
- ↑ 43.0 43.1 "The neurospora circadian system". Journal of Biological Rhythms 19 (5): 414–24. October 2004. doi:10.1177/0748730404269116. PMID 15534321.
- ↑ "The diversity and evolution of circadian clock proteins in fungi". Mycologia 102 (2): 269–78. 2010. doi:10.3852/09-073. PMID 20361495.
- ↑ "A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana". Proceedings of the National Academy of Sciences of the United States of America 112 (28): 8744–9. 2015. doi:10.1073/pnas.1508432112. PMID 26124115. Bibcode: 2015PNAS..112.8744H.
- ↑ "Analysis of Circadian Rhythms in the Basal Filamentous Ascomycete Pyronema confluens". G3: Genes, Genomes, Genetics 5 (10): 2061–71. 2015. doi:10.1534/g3.115.020461. PMID 26254031.
- ↑ Lee, SJ., Kong, M., Morse, D. Hijri, M. (2018) Expression of putative circadian clock components in the arbuscular mycorrhizal fungus Rhizoglomus irregulare. Mycorrhiza. https://doi.org/10.1007/s00572-018-0843-y
- ↑ "How fungi keep time: circadian system in Neurospora and other fungi". Current Opinion in Microbiology 9 (6): 579–87. December 2006. doi:10.1016/j.mib.2006.10.008. PMID 17064954.
- ↑ "Non-optimal codon usage affects expression, structure and function of clock protein FRQ". Nature 495 (7439): 111–5. March 2013. doi:10.1038/nature11833. PMID 23417067. Bibcode: 2013Natur.495..111Z.
- ↑ "Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ". Molecular Cell 52 (6): 832–43. December 2013. doi:10.1016/j.molcel.2013.11.005. PMID 24316221.
 |

