Biology:Acyl-protein thioesterase
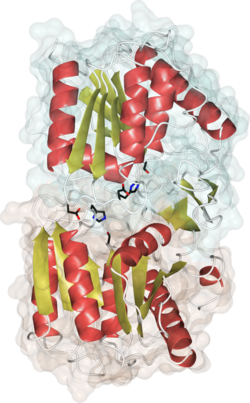 Crystal structure of human APT1, PDB code 1fj2. Alpha helices are in red, beta strands in gold, catalytic site residues in black. The 2 different monomers of the dimer are shaded in green and brown. | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | Acyl-protein thioesterases (APTs) | ||||||||
| Pfam | PF02230 | ||||||||
| InterPro | IPR029058 | ||||||||
| |||||||||
Acyl-protein thioesterases are enzymes that cleave off lipid modifications on proteins, located on the sulfur atom of cysteine residues linked via a thioester bond.[1] Acyl-protein thioesterases are part of the α/β hydrolase superfamily of proteins and have a conserved catalytic triad.[2] For that reason, acyl-protein thioesterases are also able to hydrolyze oxygen-linked ester bonds.
Function
Acyl-protein thioesterases are involved in the depalmitoylation of proteins, meaning they cleave off palmitoyl modifications on proteins' cysteine residues. Cellular targets include trimeric G-alpha proteins,[3] ion channels[4] and GAP-43.[5] Moreover, human acyl-protein thioesterases 1 and 2 have been identified as major components in controlling the palmitoylation cycle of the oncogene Ras.[6][7] Depalmitoylation of Ras by acyl-protein thioesterases potentially reduces Ras' affinity to endomembranes, allowing it to be palmitoylated again at the Golgi apparatus and to be directed to the plasma membrane. Acyl-protein thioesterases, therefore, are thought to correct potential mislocalization of Ras.
Known enzymes
|
| ||||||||||||||||||||||||||||||||||||||||||||
Currently fully validated human acyl-protein thioesterases are APT1[8] and APT2[9] which share 66% sequence homology.[10] Additionally there are a handful of putative acyl-protein thioesterases reported, including the ABHD17 enzyme family.[11][12] In the lysosome, PPT1 of the palmitoyl protein thioesterase family has similar enzymatic activity as acyl-protein thioesterases.
Structure
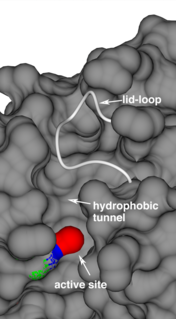
Acyl-protein thioesterases feature 3 major structural components that determine protein function and substrate processing: 1. A conserved, classical catalytic triad to break ester and thioester bonds;[2] 2. A long hydrophobic substrate tunnel to accommodate the palmitoyl moiety, as identified in the crystal structures of human acyl-protein thioesterase 1,[2] human acyl-protein thioesterase 2[13] and Zea mays acyl-protein thioesterase 2;[14] 3. A lid-loop that covers the catalytic site, is highly flexible and is a main factor determining the enzyme's product release rate.[14]

Inhibition
The involvement in controlling the localization of the oncogene Ras has made acyl-protein thioesterases potential cancer drug targets.[15] Inhibition of acyl-protein thioesterases is believed to increase mislocalization of Ras at the cell's membranes, eventually leading to a collapse of the Ras cycle. Inhibitors for acyl-protein thioesterases have been specifically targeting the hydrophobic substrate tunnel,[16][13] the catalytic site serine[17] or both.[18]
Research
Current approaches to study the biological activity of Acyl-protein Thioesterases include proteomics, monitoring the trafficking of microinjected fluorescent substrates,[19][7] the use of cell-permeable substrate mimetics,[20] and cell permeable small molecule fluorescent chemical tools.[21][22][23][24]
References
- ↑ "Protein acyl thioesterases (Review)". Molecular Membrane Biology 26 (1): 32–41. January 2009. doi:10.1080/09687680802629329. PMID 19115143.
- ↑ 2.0 2.1 2.2 "Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A". Structure 8 (11): 1137–46. November 2000. doi:10.1016/s0969-2126(00)00529-3. PMID 11080636.
- ↑ "A specific human lysophospholipase: cDNA cloning, tissue distribution and kinetic characterization". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1437 (2): 157–69. February 1999. doi:10.1016/s1388-1981(99)00012-8. PMID 10064899.
- ↑ "Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels". The Journal of Biological Chemistry 287 (18): 14718–25. April 2012. doi:10.1074/jbc.M111.335547. PMID 22399288.
- ↑ "Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43". PLOS ONE 5 (11): e15045. November 2010. doi:10.1371/journal.pone.0015045. PMID 21152083. Bibcode: 2010PLoSO...515045T.
- ↑ "An acylation cycle regulates localization and activity of palmitoylated Ras isoforms". Science 307 (5716): 1746–52. March 2005. doi:10.1126/science.1105654. PMID 15705808. Bibcode: 2005Sci...307.1746R.
- ↑ 7.0 7.1 "Small-molecule inhibition of APT1 affects Ras localization and signaling". Nature Chemical Biology 6 (6): 449–56. June 2010. doi:10.1038/nchembio.362. PMID 20418879.
- ↑ "A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS)". The Journal of Biological Chemistry 273 (25): 15830–7. June 1998. doi:10.1074/jbc.273.25.15830. PMID 9624183.
- ↑ "Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43". PLOS ONE 5 (11): e15045. November 2010. doi:10.1371/journal.pone.0015045. PMID 21152083. Bibcode: 2010PLoSO...515045T.
- ↑ "Palmitoylation and depalmitoylation dynamics at a glance". Journal of Cell Science 123 (Pt 23): 4007–10. December 2010. doi:10.1242/jcs.059287. PMID 21084560.
- ↑ "ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization". eLife 4: e11306. December 2015. doi:10.7554/eLife.11306. PMID 26701913.
- ↑ "The metabolic serine hydrolases and their functions in mammalian physiology and disease". Chemical Reviews 111 (10): 6022–63. October 2011. doi:10.1021/cr200075y. PMID 21696217.
- ↑ 13.0 13.1 "Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2)". ACS Chemical Biology 11 (12): 3374–3382. December 2016. doi:10.1021/acschembio.6b00720. PMID 27748579.
- ↑ 14.0 14.1 "A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors". Nature Communications 8 (1): 2201. December 2017. doi:10.1038/s41467-017-02347-w. PMID 29259199. Bibcode: 2017NatCo...8.2201B.
- ↑ "Targeting protein palmitoylation: selective inhibitors and implications in disease". Expert Opinion on Drug Discovery 9 (9): 1005–19. September 2014. doi:10.1517/17460441.2014.933802. PMID 24967607.
- ↑ "Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M". Angewandte Chemie 50 (42): 9838–42. October 2011. doi:10.1002/anie.201102967. PMID 21905186.
- ↑ "Boron-based inhibitors of acyl protein thioesterases 1 and 2". ChemBioChem 14 (1): 115–22. January 2013. doi:10.1002/cbic.201200571. PMID 23239555.
- ↑ "2-Bromopalmitate reduces protein deacylation by inhibition of acyl-protein thioesterase enzymatic activities". PLOS ONE 8 (10): e75232. 2013. doi:10.1371/journal.pone.0075232. PMID 24098372. Bibcode: 2013PLoSO...875232P.
- ↑ "Chemical-biological exploration of the limits of the Ras de- and repalmitoylating machinery". ChemBioChem 13 (7): 1017–23. May 2012. doi:10.1002/cbic.201200078. PMID 22488913.
- ↑ "Sensitive and rapid analysis of protein palmitoylation with a synthetic cell-permeable mimic of SRC oncoproteins". Journal of the American Chemical Society 124 (11): 2444–5. March 2002. doi:10.1021/ja017671x. PMID 11890786.
- ↑ "A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling". Nature Chemical Biology 13 (2): 150–152. February 2017. doi:10.1038/nchembio.2262. PMID 27992880.
- ↑ "A Fluorescent Probe with Improved Water Solubility Permits the Analysis of Protein S-Depalmitoylation Activity in Live Cells". Biochemistry 57 (2): 221–225. January 2018. doi:10.1021/acs.biochem.7b00835. PMID 29023093.
- ↑ "S-depalmitoylases in live cells and tissues". Chemical Science 8 (11): 7588–7592. November 2017. doi:10.1039/C7SC02805A. PMID 29568422.
- ↑ "Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes". Nature Communications 9 (1): 334. January 2018. doi:10.1038/s41467-017-02655-1. PMID 29362370. Bibcode: 2018NatCo...9..334K.
 |