Biology:Alpha-D-phosphohexomutase superfamily
| Alpha-D-phosphohexomutase | |
|---|---|
| Identifiers | |
| Symbol | ? |
| InterPro | IPR005841 |
| CDD | cd03084 |
The alpha-D-phosphohexomutases are a large superfamily of enzymes, with members in all three domains of life. Enzymes from this superfamily are ubiquitous in organisms from E. coli to humans, and catalyze a phosphoryl transfer reaction on a phosphosugar substrate. Four well studied subgroups in the superfamily are:
- Phosphoglucomutase (PGM)
- Phosphoglucomutase/Phosphomannomutase (PGM/PMM)
- Phosphoglucosamine mutase (PNGM)
- Phosphoaceytlglucosamine mutase (PAGM)
Other enzymes in the superfamily are known to act as glucose 1,6-bisphosphate synthases and phosphopentomutases.
Background
A number of proteins in the superfamily have been characterized functionally and structurally. This table illustrates different members of the superfamily.
Catalytic reaction
The enzymes in the superfamily typically catalyze the reversible conversion of 1-phosphosugars to 6-phosphosugars. The reaction proceeds via a bisphosphorylated sugar intermediate. The active form of the enzyme is phosphorylated at a conserved serine residue in the active site, and also requires a bound metal ion, typically Mg2+ for full activity. The initial phosphoryl transfer takes place from the phosphoserine to the substrate, creating a bisphosphorylated sugar intermediate. This is followed by a second phosphoryl transfer from the substrate back to enzyme, producing product and regenerating the active form of the enzyme.[36]
Structure and oligomeric state
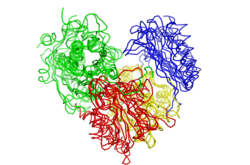
Structures of multiple enzymes have been determined through X-ray crystallography. In general, they share a very similar topology. With a heart-shape and four domains (see image below), most enzymes appear to be monomers.
However some are known to exist as dimers or tetramers in solution. Eleven crystal structures for this superfamily have been determined thus far, six of which are likely oligomers. Two distinct dimers and one tetrameric arrangement has been documented.[37]
Subgroups
There are 4 well characterized enzyme subgroups in this superfamily, which differ in their specificity for the sugar moiety of the substrate.
PGM
Phosphoglucomutase (PGM) converts D-glucose-1-phosphate into D-glucose-6-phosphate, participating in glucose breakdown & synthesis. Bacterial and eukaryotic organisms are known to have PGM enzymes, with 415 representatives currently listed in the PIR database.[38] Among bacteria, Salmonella typhimurium and Thermus thermophilus have PGM enzymes of characterized 3D structure. In eukaryotes, PGM enzymes from Oryctolagus cuniculus (rabbit) and Paramecium tetraurelia also have been structurally characterized. The highest resolution structure is from Salmonella typhimurium (1.7 A), with PDB ID 3na5. In addition, biochemical studies have shown that PGM from S. typhimurium is a dimer in solution based on analytical ultracentrifugation and small-angle X-ray scattering (SAXS). [39]
PMM/PGM
Phosphoglucomutase/phosphomannomutase (PGM/PMM)- Enzymes from this subgroup can use either mannose or glucose-based phosphosugar substrates with equal efficiency. PMM/PGM enzymes are found mainly in bacterial organisms, with a total of 1,331 representatives currently listed in the PIR database.[40] These enzymes are involved in the biosynthesis of many different carbohydrates and glycolipids, which vary depending on the organism. The best studied enzyme from this subgroup is from the bacterium, Pseudomonas aeruginosa, where PMM/PGM participate in multiple biosynthetic pathways including those of lipopolysaccharide, alginate and rhamnolipid.
Structural studies of P. aeruginosa PMM/PGM by X-ray crystallography have been conducted as both apo-enzyme and as protein-ligand complexes. Based on these studies, it has been seen that when the sugar substrate binds to the enzyme there is a rotation in the C-terminal domain of the protein. This changes the active site from an open cleft in the apo-enzyme into a nearly solvent inaccessible pocket. This theme of conformational flexibility, particularly with regard to the C-terminal domain of these enzymes, has been observed in multiple proteins in the superfamily.
PNGM
Phosphoglucosamine mutase (PNGM) participates in the biosynthesis of UDP-N-acetylglucosamine (UDP-GlcNAc). This bacterial enzyme has been conserved throughout evolution and is involved in the cytoplasmic steps of peptidoglycan biosynthesis, which is essential for bacterial survival and is also not present in humans.[41]
PAGM
Phosphoacetylglucosamine mutase (PAGM)- To date, this subgroup contains 178 members, with all known being eukaryotic.[42] There is only one known organism with known structure, it is Candida albicans. Like PNGM, it is involved in the biosynthesis of UDP-N-acetylglucosamine. UDP-GlcNAc is a UDP sugar that works as a biosynthetic precursor of glycoproteins, mucopolysaccharides, and the cell wall of bacteria. AGM1, a characterized structure of PAGM, catalyzes the conversion of N-acetylglucosamine 6-phosphate to N-acetylglucosamine 1-phosphate. AGM1 structure was determined from Candida albicans in the apoform and complex forms with substrate and product. Like other enzymes in the superfamily, it has four domains, with two additional beta-strands in domain four and a circular permutation in domain 1.[43]
References
- ↑ "Lipooligosaccharide biosynthesis in pathogenic Neisseria. Cloning, identification, and characterization of the phosphoglucomutase gene". The Journal of Biological Chemistry 269 (15): 11162–9. April 1994. doi:10.1016/S0021-9258(19)78105-8. PMID 8157643.
- ↑ "Role of phosphoglucomutase in lipooligosaccharide biosynthesis in Neisseria gonorrhoeae". Journal of Bacteriology 176 (10): 2930–7. May 1994. doi:10.1128/jb.176.10.2930-2937.1994. PMID 8188595.
- ↑ "Phase-variation of the truncated lipo-oligosaccharide of Neisseria meningitidis NMB phosphoglucomutase isogenic mutant NMB-R6". Carbohydrate Research 338 (24): 2905–12. November 2003. doi:10.1016/j.carres.2003.08.014. PMID 14667712.
- ↑ "Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus". Journal of Bacteriology 189 (6): 2521–30. March 2007. doi:10.1128/jb.01683-06. PMID 17209021.
- ↑ "The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase". Journal of Bacteriology 179 (17): 5321–5. September 1997. doi:10.1128/jb.179.17.5321-5325.1997. PMID 9286983.
- ↑ "Effects of alpha-phosphoglucomutase deficiency on cell wall properties and fitness in Streptococcus gordonii". Microbiology 153 (Pt 2): 490–8. February 2007. doi:10.1099/mic.0.29256-0. PMID 17259620.
- ↑ "Identification of the Streptococcus gordonii glmM gene encoding phosphoglucosamine mutase and its role in bacterial cell morphology, biofilm formation, and sensitivity to antibiotics". FEMS Immunology and Medical Microbiology 53 (2): 166–77. July 2008. doi:10.1111/j.1574-695X.2008.00410.x. PMID 18462386.
- ↑ "Contribution of phosphoglucosamine mutase to the resistance of Streptococcus gordonii DL1 to polymorphonuclear leukocyte killing". FEMS Microbiology Letters 297 (2): 196–202. August 2009. doi:10.1111/j.1574-6968.2009.01673.x. PMID 19552711.
- ↑ "Contribution of phosphoglucosamine mutase to determination of bacterial cell morphology in Streptococcus gordonii". Odontology 100 (1): 28–33. January 2012. doi:10.1007/s10266-011-0026-1. PMID 21567120.
- ↑ "Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development". Infection and Immunity 73 (10): 6935–44. October 2005. doi:10.1128/iai.73.10.6935-6944.2005. PMID 16177373.
- ↑ "Role of phosphoglucosamine mutase on virulence properties of Streptococcus mutans". Oral Microbiology and Immunology 24 (4): 272–7. August 2009. doi:10.1111/j.1399-302x.2009.00503.x. PMID 19572887.
- ↑ "Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase". Journal of Bacteriology 182 (7): 1854–63. April 2000. doi:10.1128/jb.182.7.1854-1863.2000. PMID 10714989.
- ↑ "Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae". Infection and Immunity 69 (4): 2309–17. April 2001. doi:10.1128/iai.69.4.2309-2317.2001. PMID 11254588.
- ↑ "Overexpression of Mycobacterium tuberculosis manB, a phosphomannomutase that increases phosphatidylinositol mannoside biosynthesis in Mycobacterium smegmatis and mycobacterial association with human macrophages". Molecular Microbiology 58 (3): 774–90. November 2005. doi:10.1111/j.1365-2958.2005.04862.x. PMID 16238626.
- ↑ "Identification of M. tuberculosis Rv3441c and M. smegmatis MSMEG_1556 and essentiality of M. smegmatis MSMEG_1556". PLOS ONE 7 (8): e42769. 2012. doi:10.1371/journal.pone.0042769. PMID 22905172. Bibcode: 2012PLoSO...742769L.
- ↑ "Role of phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence". Infection and Immunity 68 (8): 4673–80. August 2000. doi:10.1128/iai.68.8.4673-4680.2000. PMID 10899872.
- ↑ "Brucella abortus rough mutants are cytopathic for macrophages in culture". Infection and Immunity 72 (1): 440–50. January 2004. doi:10.1128/iai.72.1.440-450.2004. PMID 14688125.
- ↑ "Evaluation of Brucella abortus phosphoglucomutase (pgm) mutant as a new live rough-phenotype vaccine". Infection and Immunity 71 (11): 6264–9. November 2003. doi:10.1128/iai.71.11.6264-6269.2003. PMID 14573645.
- ↑ "Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication". Infection and Immunity 68 (10): 5716–23. October 2000. doi:10.1128/iai.68.10.5716-5723.2000. PMID 10992476.
- ↑ "Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen". Infection and Immunity 77 (10): 4197–208. October 2009. doi:10.1128/iai.00417-09. PMID 19651862.
- ↑ "Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli". Journal of Bacteriology 176 (18): 5847–51. September 1994. doi:10.1128/jb.176.18.5847-5851.1994. PMID 8083177.
- ↑ "Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli". The Journal of Biological Chemistry 271 (1): 32–9. January 1996. doi:10.1074/jbc.271.1.32. PMID 8550580.
- ↑ "The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis". Molecular Microbiology 48 (3): 737–51. May 2003. doi:10.1046/j.1365-2958.2003.03450.x. PMID 12694618.
- ↑ "Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor". Molecular Microbiology 37 (1): 13–27. July 2000. doi:10.1046/j.1365-2958.2000.01952.x. PMID 10931302.
- ↑ "The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase". Journal of Bacteriology 179 (11): 3488–93. June 1997. doi:10.1128/jb.179.11.3488-3493.1997. PMID 9171391.
- ↑ "Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection". Infection and Immunity 63 (10): 4166–9. October 1995. doi:10.1128/iai.63.10.4166-4169.1995. PMID 7558335.
- ↑ "Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection". Infection and Immunity 64 (1): 37–43. January 1996. doi:10.1128/iai.64.1.37-43.1996. PMID 8557368.
- ↑ "Chromosomal organization and transcription analysis of genes in the vicinity of Pseudomonas aeruginosa glmM gene encoding phosphoglucosamine mutase". Biochemical and Biophysical Research Communications 302 (2): 363–71. March 2003. doi:10.1016/s0006-291x(03)00169-4. PMID 12604356.
- ↑ "The enzyme phosphoglucomutase (Pgm) is required by Salmonella enterica serovar Typhimurium for O-antigen production, resistance to antimicrobial peptides and in vivo fitness". Microbiology 155 (Pt 10): 3403–10. October 2009. doi:10.1099/mic.0.029553-0. PMID 19589833.
- ↑ "Genome subtraction for novel target definition in Salmonella typhi". Bioinformation 4 (4): 143–50. October 2009. doi:10.6026/97320630004143. PMID 20198190.
- ↑ "Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance". Infection and Immunity 71 (6): 3068–75. June 2003. doi:10.1128/iai.71.6.3068-3075.2003. PMID 12761084.
- ↑ "Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation". International Journal of Antimicrobial Agents 35 (2): 126–30. February 2010. doi:10.1016/j.ijantimicag.2009.09.015. PMID 19926255. http://ntur.lib.ntu.edu.tw//handle/246246/189289.
- ↑ "Genetic analysis of phosphomannomutase/phosphoglucomutase from Vibrio furnissii and characterization of its role in virulence". Archives of Microbiology 180 (4): 240–50. October 2003. doi:10.1007/s00203-003-0582-z. PMID 12904831.
- ↑ "rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization". Infection and Immunity 67 (2): 976–80. February 1999. doi:10.1128/IAI.67.2.976-980.1999. PMID 9916119.
- ↑ "Phosphoglucomutase of Yersinia pestis is required for autoaggregation and polymyxin B resistance". Infection and Immunity 78 (3): 1163–75. March 2010. doi:10.1128/iai.00997-09. PMID 20028810.
- ↑ "Evolutionary trace analysis of the alpha-D-phosphohexomutase superfamily". Protein Science 13 (8): 2130–8. August 2004. doi:10.1110/ps.04801104. PMID 15238632.
- ↑ "Conservation of functionally important global motions in an enzyme superfamily across varying quaternary structures". Journal of Molecular Biology 423 (5): 831–46. November 2012. doi:10.1016/j.jmb.2012.08.013. PMID 22935436.
- ↑ "Summary Report for PIRSF001493". http://pir.georgetown.edu/cgi-bin/ipcSF?id=PIRSF001493.
- ↑ "Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens". Proteins 79 (4): 1215–29. April 2011. doi:10.1002/prot.22957. PMID 21246636.
- ↑ "Summary Report for PIRSF005849". http://pir.georgetown.edu/cgi-bin/ipcSF?id=PIRSF005849.
- ↑ "Crystal structure of Bacillus anthracis phosphoglucosamine mutase, an enzyme in the peptidoglycan biosynthetic pathway". Journal of Bacteriology 193 (16): 4081–7. August 2011. doi:10.1128/jb.00418-11. PMID 21685296.
- ↑ "Summary report for PIRSF016408". http://pir.georgetown.edu/cgi-bin/ipcSF?id=PIRSF016408.
- ↑ "Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, and its substrate and product complexes". The Journal of Biological Chemistry 281 (28): 19740–7. July 2006. doi:10.1074/jbc.m600801200. PMID 16651269.
 |