Biology:Ced-9
| CED-9 (gene) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | CED-9 | ||||||
| Entrez | 3565776 | ||||||
| HomoloGene | 80344 | ||||||
| PDB | 1OHU (ECOD) | ||||||
| RefSeq (mRNA) | NM_066883 | ||||||
| RefSeq (Prot) | NP_499284 | ||||||
| UniProt | P41958 | ||||||
| Other data | |||||||
| Chromosome | III: 10.34 - 10.34 Mb | ||||||
| |||||||
Cell death abnormality gene 9 (CED-9), also known as apoptosis regulator CED-9, is a gene found in Caenorhabditis elegans that inhibits/represses programmed cell death (apoptosis).[1] The gene was discovered while searching for mutations in the apoptotic pathway after the discovery of the apoptosis promoting genes CED-3 and CED-4.[2] The gene gives rise to the apoptosis regulator CED-9 protein found as an Integral membrane protein in the mitochondrial membrane.[3] The protein is homologous to the human apoptotic regulator Bcl-2 as well as all other proteins in the Bcl-2 protein family.[4] CED-9 is involved in the inhibition of CED-4 which is the activator of the CED-3 caspase.[5] Because of the pathway homology with humans as well as the specific protein homology, CED-9 has been used to represent the human cell apoptosis interactions of Bcl-2 in research.[6]
Discovery
The CED-9 gene was discovered in 1992 while searching the genome of C. elegans for mutations affecting cell death.[2] The first mutation identified was a dominant gain of function mutation referred to as n1950 that allowed cells to survive when they were fated to die.[2] The observed phenotype was similar to that observed in CED-3 and CED-4 loss of function mutants (known proteins from the apoptotic pathway).[2] It was also observed that loss of function mutations in CED-3 and CED-4 were able to rescue cells with a CED-9 loss of function mutation.[7] These observations suggested that CED-9 functioned upstream of the CED-3 and CED-4 proteins in the same pathway.[7]
Structure
Gene
The CED-9 gene is located on chromosome 3 of the C. elegans genome.[1] CED-9 is transcribed from a polycistronic locus that also contains genes required for the mitochondrial Oxidative phosphorylation.[4] The CED-9 gene has been identified in two distinct transcripts, both transcribed from this locus.[4] The first was identified as a 1.3 kb transcript encoding only the CED-9 sequence. The second being a rare 2.1 kb bicistronic transcript containing the 1.3 kb transcript and an additional 0.75 kb transcript from an upstream gene found in the gene locus.[4] this 0.75 kb transcript corresponds to the cytochrome protein cyt-1 that functions in the electron transport chain within the mitochondria.[4] The bicistronic transcript is then spliced giving rise to the two distinct mature messenger RNA (mRNA) for both genes.[4] The most prevalent transcript however, is the 1299-nucleotide (~1.3kb) long transcript that encodes an 843-nucleotide mRNA containing 4 exons.[8]
Protein
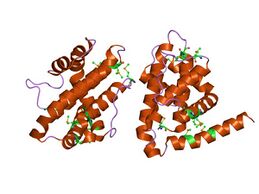
CED-9 encodes the apoptosis regulator CED-9 protein which is an important negative regulator protein in the apoptosis pathway of C. elegans.[9] The protein consists of 280 amino acids and has a molecular weight of 31824.42 Da.[10] The structure of this protein has been solved using X-ray diffraction revealing 9 Helices, 2 Beta strands, and 2 turn motifs.[3] The CED-9 protein belongs to the Bcl-2-like protein family. This refers to the homology between the ced-9 protein and the B-cell lymphoma proteins (Bcl) found in humans, specifically the Bcl-2 protein.[4] CED-9 contains a BCL domain homologous to Bcl-2 domains BH1, BH2, and part of BH3 as well as a separate domain homologous to BH4 located near the N-terminus.[11] CED-9 also includes a transmembrane domain on the C-terminal end of the structure that anchors the protein to the mitochondrial membrane.[12] However, research shown that the C-terminal domain is not necessary for the protein's main function as an inhibitor of the CED-4 protein found in the same apoptosis signalling pathway.[12]
Function
Cell death, or apoptosis during early development is crucial for the correct morphology and refractivity of adult C. elegans.[13] This process involves a signal and interaction cascade of proteins leading to the engulfment and death of the targeted cell. Proteins in this cascade can be categorized into two groups; pro-apoptotic and anti-apoptotic.[13] Pro-apoptotic proteins activate the apoptosis pathway while anti-apoptotic proteins suppress the pathway.[14] CED-9 is classified as an anti-apoptotic protein.[15]
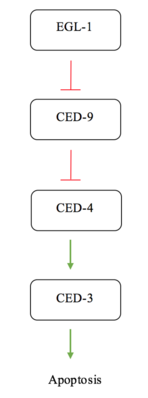
Apoptosis in C. elegans is often simplified to interactions between four major proteins in the pathway; EGL-1, CED-9, CED-4, and CED-3. CED-3 is the final protease in the interaction network and is responsible for activating the proteins involved in cell disassembly.[13] CED-9 is said to protect cells from the apoptosis pathway.[7] Under normal conditions, in a cell not experiencing apoptotic signalling, CED-9 forms a complex with CED-4 at the membrane of the mitochondria.[14] This interaction sequesters the pro-apoptotic signalling of CED-4. CED-4 consists of an asymmetric dimer of CED-4a and CED-4b proteins in which CED-9 can specifically recognize and bind CED-4a.[14] This interaction is a highly specific recognition and binding interaction between the N-terminal tails of both proteins.[14] When the cell receives an apoptotic signal via a receptor commonly referred to as a "death receptor", the protein EGL-1 is activated.[5] The active EGL-1 binds CED-9 causing a Conformational change that interrupts and inhibits the CED-9 - CED-4 interaction.[14] CED-4 is free to dissociate and activate the CED-3 protease effectively triggering the final stages for apoptosis.[5]
Mutations
The cells developed during embryogenesis and early life in C. elegans have one of two fates, to live and differentiate or apoptose.[16] Apoptosis during development is highly regulated and only occurs in specific cells at specific times.[16] Every cell division and cell death in the development of C. elegans from embryo to adult has been studied and documented to reveal a fixed pattern between individual organisms.[16] Apoptosis during development is important for the proper morphology and refractivity of C. elegans, but it is not always essential for survival.[16] Thus, over 100 mutations have been observed and documented as affecting the apoptotic pathway of C. elegans.[7] Many proteins involved in the interaction cascade were discovered because of these mutations and their resulting phenotype. CED-9 mutants are among the mutations that affect this pathway. CED-9 gain of function mutations are unresponsive to apoptosis signalling and allow cells fated to die, to survive.[7] A notable example of a CED-9 dominant gain of function mutation would be the n1950 mutation which was the first mutation documented for CED-9 and responsible for the gene's discovery.[2] Loss of function mutations cause inappropriate cell death in the absence of apoptosis stimuli.[7] Mutations in CED-9 also reveal its maternal effect; where the genotype of the mother determines the phenotype of the progeny.[7] Homozygous, loss of function mutants from a heterozygous mother experience some unpredictable cell death, however, give rise to unviable progeny themselves.[7]
Significance
The apoptotic pathway has been conserved in evolutionary history and is vital for the maintenance of multicellular organisms such as humans.[17] A parallel pathway to the one found in C. elegans is also observed in mammals involving a number of homologous proteins. Disruptions to this pathway often lead to diseases that, in humans, include various cancers, autoimmune diseases, and neurodegenerative disease.[17] Bcl-2 in particular is often found mutated in many human cancers.[6] Due to the conserved nature of the apoptotic pathway and the extensive knowledge and understanding available for C. elegans, the organisms apoptotic pathway can be used as a proxy for the human equivalent.[17] CED-9 is the homologue of Bcl-2 which can provide researchers with information including the pathways the protein is involved in and the consequences of mutation that may parallel pathways or abnormalities in humans.[6]
Interactions
References
- ↑ 1.0 1.1 "ced-9 Apoptosis regulator ced-9 [Caenorhabditis elegans - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/?term=3565776.
- ↑ 2.0 2.1 2.2 2.3 2.4 "Caenorhabditis elegans gene ced-9 protects cells from programmed cell death". Nature 356 (6369): 494–9. April 1992. doi:10.1038/356494a0. PMID 1560823. Bibcode: 1992Natur.356..494H.
- ↑ 3.0 3.1 "ced-9 - Apoptosis regulator ced-9 - Caenorhabditis elegans - ced-9 gene & protein" (in en). https://www.uniprot.org/uniprot/P41958.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Hengartner, Michael O.; Horvitz, H. Robert (1994-02-25). "C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2". Cell 76 (4): 665–676. doi:10.1016/0092-8674(94)90506-1. PMID 7907274.
- ↑ 5.0 5.1 5.2 "Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation". The Journal of Biological Chemistry 273 (50): 33495–500. December 1998. doi:10.1074/jbc.273.50.33495. PMID 9837929.
- ↑ 6.0 6.1 6.2 "Apoptosis in C. elegans: lessons for cancer and immunity". Frontiers in Cellular and Infection Microbiology 3: 67. 2013-10-18. doi:10.3389/fcimb.2013.00067. PMID 24151577.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 Riddle, Donald L.; Blumenthal, Thomas; Meyer, Barbara J.; Priess, James R. (1997) (in en). Genetics of Programmed Cell Death. Cold Spring Harbor Laboratory Press. https://www.ncbi.nlm.nih.gov/books/NBK20015/.
- ↑ 8.0 8.1 8.2 "ced-9". http://www.wormbase.org/species/c_elegans/gene/WBGene00000423#04-9g-3.
- ↑ "Family: BH4 (PF02180)". http://pfam.xfam.org/family/BH4.
- ↑ "ExPASy" (in en-US). http://web.expasy.org/cgi-bin/protparam/protparam1?P41958@1-280@.
- ↑ "SMART: Sequence analysis results for CED9_CAEEL" (in en). https://smart.embl.de/smart/show_motifs.pl?ID=P41958.
- ↑ 12.0 12.1 "Regulation of apoptosis by C. elegans CED-9 in the absence of the C-terminal transmembrane domain". Cell Death and Differentiation 14 (11): 1925–35. November 2007. doi:10.1038/sj.cdd.4402215. PMID 17703231.
- ↑ 13.0 13.1 13.2 "Programmed Cell Death During Caenorhabditis elegans Development". Genetics 203 (4): 1533–62. August 2016. doi:10.1534/genetics.115.186247. PMID 27516615.
- ↑ 14.0 14.1 14.2 14.3 14.4 "Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans". Nature 437 (7060): 831–7. October 2005. doi:10.1038/nature04002. PMID 16208361. Bibcode: 2005Natur.437..831Y.
- ↑ "Anti-apoptotic versus pro-apoptotic signal transduction: checkpoints and stop signs along the road to death". Oncogene 17 (11 Reviews): 1475–82. September 1998. doi:10.1038/sj.onc.1202183. PMID 9779994.
- ↑ 16.0 16.1 16.2 16.3 "Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans". Science 220 (4603): 1277–9. June 1983. doi:10.1126/science.6857247. PMID 6857247. Bibcode: 1983Sci...220.1277H.
- ↑ 17.0 17.1 17.2 "Apoptosis and disease: regulation and clinical relevance of programmed cell death". Annual Review of Medicine 48: 267–81. 1997. doi:10.1146/annurev.med.48.1.267. PMID 9046961.
- ↑ 18.0 18.1 18.2 Lab, Mike Tyers. "ced-9 (T07C4.8) Result Summary | BioGRID" (in en). https://thebiogrid.org/533094.
 |