Biology:Cyanobacterial clock proteins
| KaiA domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
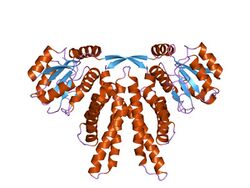 crystal structure of circadian clock protein kaia from synechococcus elongatus | |||||||||
| Identifiers | |||||||||
| Symbol | KaiA | ||||||||
| Pfam | PF07688 | ||||||||
| InterPro | IPR011648 | ||||||||
| |||||||||
| KaiB domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
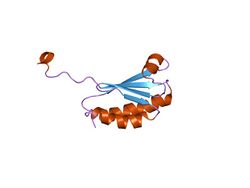 solution structure of the n-terminal domain of synechococcus elongatus sasa (average minimized structure) | |||||||||
| Identifiers | |||||||||
| Symbol | KaiB | ||||||||
| Pfam | PF07689 | ||||||||
| Pfam clan | CL0172 | ||||||||
| InterPro | IPR011649 | ||||||||
| CDD | cd02978 | ||||||||
| |||||||||
| KaiC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
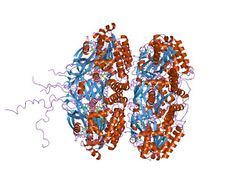 crystal structure of full length circadian clock protein kaic with phosphorylation sites | |||||||||
| Identifiers | |||||||||
| Symbol | KaiC | ||||||||
| Pfam | PF06745 | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR014774 | ||||||||
| CDD | cd01124 | ||||||||
| |||||||||
In molecular biology, the cyanobacterial clock proteins are the main circadian regulator in cyanobacteria. The cyanobacterial clock proteins comprise three proteins: KaiA, KaiB and KaiC. The kaiABC complex may act as a promoter-nonspecific transcription regulator that represses transcription, possibly by acting on the state of chromosome compaction. This complex is expressed from a KaiABC operon.
See also: bacterial circadian rhythms
In the complex, KaiA enhances the phosphorylation status of kaiC. In contrast, the presence of kaiB in the complex decreases the phosphorylation status of kaiC, suggesting that kaiB acts by antagonising the interaction between kaiA and kaiC. The activity of KaiA activates kaiBC expression, while KaiC represses it.
Also in the KaiC family is RadA/Sms, a highly conserved eubacterial protein that shares sequence similarity with both RecA strand transferase and lon protease. The RadA/Sms family are probable ATP-dependent proteases involved in both DNA repair and degradation of proteins, peptides, glycopeptides. They are classified in as non-peptidase homologues and unassigned peptidases in MEROPS peptidase family S16 (lon protease family, clan SJ). RadA/Sms is involved in recombination and recombinational repair, most likely involving the stabilisation or processing of branched DNA molecules or blocked replication forks because of its genetic redundancy with RecG and RuvABC.[1]
Structure
The overall fold of the KaiA monomer is that of a four-helix bundle, which forms a dimer in the known structure.[2] KaiA functions as a homodimer. Each monomer is composed of three functional domains: the N-terminal amplitude-amplifier domain, the central period-adjuster domain and the C-terminal clock-oscillator domain. The N-terminal domain of KaiA, from cyanobacteria, acts as a pseudo-receiver domain, but lacks the conserved aspartyl residue required for phosphotransfer in response regulators.[3] The C-terminal domain is responsible for dimer formation, binding to KaiC, enhancing KaiC phosphorylation and generating the circadian oscillations.[4] The KaiA protein from Anabaena sp. (strain PCC 7120) lacks the N-terminal CheY-like domain.
KaiB adopts an alpha-beta meander motif and is found to be a dimer or a tetramer.[2][5]
KaiC belongs to a larger family of proteins; it performs autophosphorylation and acts as its own transcriptional repressor. It binds ATP.[6]
Alternate arrangements
This section is missing information about paralogs (KaiB2C2, KaiB3C3) [PMID 12604787]; alternate KaiA in the form of KaiA3 [10.1101/2021.07.20.453058]; systems without KaiA. (September 2021) |
History of discovery
Due to the lack of a nucleus in these organisms, there was doubt as to whether or not cyanobacteria would be able to express circadian rhythms. Kondo et al. were the first to definitively demonstrate that cyanobacteria do in fact have circadian rhythms. In a 1993 experiment, they used a luciferase reporter inserted into the genetically tractable Synechococcus sp., which was grown in a 12:12 light-dark cycle to ensure “entrainment”. There were two sets of bacteria so that one was in light while the other was in darkness during this entrainment period. Once the bacteria entered the stationary phase, they were transferred into test tubes kept in constant light, except for 5-minute recording periods every 30 minutes, in which the tubes were kept in darkness to measure their levels of bioluminescence. They found that the level of bioluminescence cycled at a near 24-hour period, and that the two groups oscillated with opposite phases. This led them to conclude that the Synechococcus sp. genome was regulated by a circadian clock. (1)
Function in vitro
The circadian oscillators in eukaryotes that have been studied function using a negative feedback loop in which proteins inhibit their own transcription in a cycle that takes approximately 24 hours. This is known as a transcription-translation-derived oscillator (TTO).(2) Without a nucleus, prokaryotic cells must have a different mechanism of keeping circadian time. In 1998, Ishiura et al. determined that the KaiABC protein complex was responsible for the circadian negative feedback loop in Synechococcus by mapping 19 clock mutants to the genes for these three proteins.(3) An experiment by Nakajima et al., in 2005, was able to demonstrate the circadian oscillation of the Synechococcus KaiABC complex in vitro. They did this by adding KaiA, KaiB, KaiC, and ATP into a test tube in the approximate ratio recorded in vivo. They then measured the levels of KaiC phosphorylation and found that it demonstrated circadian rhythmicity for three cycles without damping. This cycle was also temperature compensating. They also tested incubating mutant KaiC protein with KaiA, KaiB, and ATP. They found that the period of KaiC phosphorylation matched the intrinsic period of the cyanobacterium with the corresponding mutant genome. These results led them to conclude that KaiC phosphorylation is the basis for circadian rhythm generation in Synechococcus. (2)
Cyanobacterial clocks as model systems
Cyanobacteria are the simplest organisms that have been observed demonstrating circadian rhythms.(2)(3) The primitiveness and simplicity make the KaiC phosphorylation model invaluable to circadian rhythm research. While it is much simpler than models for eukaryotic circadian rhythm generators, the principles are largely the same. In both systems the circadian period is dependent on the interactions between proteins within the cell, and when the genes for those proteins are mutated, the expressed period changes. (1)(2) This model of circadian rhythm generation also has implications for the study of circadian “evolutionary biology”. Given the simplicity of cyanobacteria and of this circadian system, it may be safe to assume that eukaryotic circadian oscillators are derived from a system similar to that present in cyanobacterium. (1)
References
- ↑ "Role for radA/sms in recombination intermediate processing in Escherichia coli". J. Bacteriol. 184 (24): 6836–44. December 2002. doi:10.1128/jb.184.24.6836-6844.2002. PMID 12446634.
- ↑ 2.0 2.1 "Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC". EMBO J. 23 (8): 1688–98. April 2004. doi:10.1038/sj.emboj.7600190. PMID 15071498.
- ↑ "Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism". Proc. Natl. Acad. Sci. U.S.A. 99 (24): 15357–62. November 2002. doi:10.1073/pnas.232517099. PMID 12438647.
- ↑ "Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein". Nat. Struct. Mol. Biol. 11 (7): 623–31. July 2004. doi:10.1038/nsmb781. PMID 15170179.
- ↑ "Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.". J Biol Chem 280 (19): 19127–35. 2005. doi:10.1074/jbc.M411284200. PMID 15716274.
- ↑ "Visualizing a circadian clock protein: crystal structure of KaiC and functional insights.". Mol Cell 15 (3): 375–88. 2004. doi:10.1016/j.molcel.2004.07.013. PMID 15304218.
 |

