Biology:Cytochrome d
| Ubiquinol oxidase (electrogenic, proton-motive force generating; Cytochrome bd) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Predicted structure of E. coli Cytochrome bd-1 | |||||||||
| Identifiers | |||||||||
| EC number | 7.1.1.7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Cytochrome d, previously known as cytochrome a2, is a name for all cytochromes (electron-transporting heme proteins) that contain heme D as a cofactor. Two unrelated classes of cytochrome d are known: Cytochrome bd, an enzyme that generates a charge across the membrane so that protons will move,[1] and cytochrome cd1 (NirS; SCOP b.70.2), a nitrite reductase.[2]
Cytochrome bd is found in plenty of aerobic bacteria, especially when it has grown with a limited oxygen supply. Compared to other terminal oxidases, it is notable for its high oxygen affinity and resistance to cyanide poisoning. It has a group of very similar relatives that do not use heme D, known as cyanide insensitive oxidases (CIOs).[3]
Function

Cytochrome d is, as other proteins of its family, a membrane-bound hemeprotein, but unlike cytochromes a and b, cytochrome D has a heme D instead of a heme A or heme B group.[4]
Cytochrome d is part of the cytochrome bd terminal oxidase which catalyse the two electron oxidation of ubiquinol. This process is an oxidative phosphorylation that oxidizes the ubiquinol-8 to ubiquinone. The chemical reaction followed by this process is:
- Ubiquinol-8 + O2 → Ubiquinone-8 + H2O[5]
By a similar reaction, it also catalyses the reduction of oxygen to water, which involves 4 electrons.
As a terminal oxidase, the reaction generates a proton motive force:
- 2 ubiquinol[inner membrane] + O2 + 4 H+[cytoplasm] → 2 ubiquinone[inner membrane] + 2 H2O + 4 H+[periplasm]
Some members of the family may accept or prefer other electron-transporting quinols such as menaquinol or plastoquinol in lieu of ubiquinol.[3]
Structure
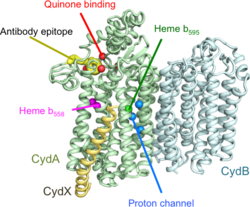
Cytochrome bd (OPM family 805) is a tri-heme oxidase as it is compound by cytochromes b558, b595 and d. Its main function is the reduction of O2 to H2O. It is thought that it uses a di-heme active site, which is formed by the hemes of cytochromes b595 and d. These two cytochromes are considered high-spin complexes, what is directly related to the electrons' spin. While other respiratory terminal oxidases which catalyze that same reaction have a heme-copper active site and use a proton pump, cytochrome bd has an active site with iron instead of copper and need no proton pump as they can produce a proton-motion force themselves.[6] They are embedded in the bacterial cytoplasmic bilayer and serve as terminal oxidases in the respiratory chain.[7]
The oxidases tend to have two or three subunits. Subunits 1 (InterPro: IPR003317) and 2 (InterPro: IPR002585) are predicted to have pseudo-symmetry, and are sufficient to bind the two heme b molecules.[8] Some proteobacterial assemblies require a third subunit (InterPro: IPR012994) to bind heme d; others do not.[9]
The high-resolution structure heterotrimeric Cytochromes bd from Geobacillus species has been determined (PDB: 5IR6, 5DOQ). The third subunit does not share sequence homology with the third subunit proteobacteria, but does come into the assemblies at a similar position.[10]
Occurrence
Escherichia coli
E. coli possess two sets of Cytochrome bd.[7] The bd-I complex (CydABX) is a heterotrimer, while the bd-II complex (AppCB) is a heterodimer. There is an AppX gene that may correspond to a subunit 3 for AppCB.[9]
The ability of bd-II to generate a proton motive force is a matter of recent debate, putting it under the nonelectrogenic Ubiquinol oxidase (H+-transporting) in some categorizations.[11]
Azotobacter vinelandii
Azotobacter vinelandii is a nitrogen-fixing bacteria which is known by its high respiratory rate among aerobic organisms. Some physiological studies postulate that cytochrome d functions as a terminal oxidase in the membranes of this organism, taking part in the electron transport system. The studies characterized the different genes in the two subunits (Q09049, C1DEL1; third subunit C1DEL0). A very extensive homology with CydAB of the E. coli was found in these studies.[12]
Spectra
Generally, in protein complexes, cytochrome D gives an absorption band of approximately 636 nm or 638 nm, depending on the cytochrome d form. If it is oxidized, the band has a length of 636 nm, and a 638 nm length if it is reduced. It is commonly associated to certain prosthetic groups when found in multiple subunit complexes. Detecting cytochrome d as Fe(II) pyridine alkaline hemachrome is very difficult because the stability under these conditions is limited. If cytochrome d is pulled out of the protein complex (as heme D) and placed in ether containing from 1 to 5 % of HCl, it gives a different absorption band (603 nm, in the oxidized form).[2]
References
- ↑ EC 7.1.1.7
- ↑ 2.0 2.1 "Nomenclature Committee of the International Union of Biochemistry (NC-IUB). Nomenclature of electron-transfer proteins. Recommendations 1989". European Journal of Biochemistry 200 (3): 599–611. September 1991. doi:10.1111/j.1432-1033.1991.tb16223.x. PMID 1655423. http://www.jbc.org/content/267/1/665.full.pdf.
- ↑ 3.0 3.1 "The cytochrome bd respiratory oxygen reductases". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1807 (11): 1398–413. November 2011. doi:10.1016/j.bbabio.2011.06.016. PMID 21756872.
- ↑ "Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability". FEBS Letters 579 (21): 4567–70. August 2005. doi:10.1016/j.febslet.2005.07.011. PMID 16087180.
- ↑ Cytochrome d ubiquinol oxidase subunit 1
- ↑ "Accommodation of CO in the di-heme active site of cytochrome bd terminal oxidase from Escherichia coli". Journal of Inorganic Biochemistry 118: 65–7. January 2013. doi:10.1016/j.jinorgbio.2012.09.016. PMID 23123340.
- ↑ 7.0 7.1 Michael J. Miller, Robert B. Gennis. The Cytochrome d Complex Is a Coupling Site in the Aerobic Respiratory Chain of Escherichia coli. The Journal of Biological Chemistry Vol.260 No.26 (1985)
- ↑ "Large-scale determination of previously unsolved protein structures using evolutionary information". eLife 4: e09248. September 2015. doi:10.7554/eLife.09248. PMID 26335199.
- ↑ 9.0 9.1 Escherichia coli K-12 substr. MG1655 Transporter: cytochrome bd-I terminal oxidase
- ↑ "Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases". Science 352 (6285): 583–6. April 2016. doi:10.1126/science.aaf2477. PMID 27126043. Bibcode: 2016Sci...352..583S.
- ↑ "Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode". Proceedings of the National Academy of Sciences of the United States of America 108 (42): 17320–4. October 2011. doi:10.1073/pnas.1108217108. PMID 21987791. Bibcode: 2011PNAS..10817320B.
- ↑ Jones CW, Redfearn ER. The cytochrome system of Azotobacter vinelandii. Biochim Biophys Acta. 1967 Sep 6;143(2):340–353
 |
