Biology:Electrophoretic color marker


An electrophoretic color marker is a chemical used to monitor the progress of agarose gel electrophoresis and polyacrylamide gel electrophoresis (PAGE) since DNA, RNA, and most proteins are colourless.[1] The color markers are made up of a mixture of dyes that migrate through the gel matrix alongside the sample of interest. They are typically designed to have different mobilities from the sample components and to generate colored bands that can be used to assess the migration and separation of sample components.[2]
Color markers are often used as molecular weight standards, loading dyes, tracking dyes, or staining solutions. Molecular weight ladders are used to estimate the size of DNA and protein fragments by comparing their migration distance to that of the colored bands.[2] DNA and protein standards are available commercially in a wide range of sizes, and are often provided with pre-stained or color-coded bands for easy identification. Loading dyes are usually added to the sample buffer before loading the sample onto the gel, and they migrate through the gel along with the sample to help track its progress during electrophoresis.[3] Tracking dyes are added to the electrophoresis buffer rather to provide a visual marker of the buffer front. Staining solutions are applied after electrophoresis to visualize the sample bands, and are available in a range of colors.
Different types of electrophoretic color markers are available commercially, with varying numbers and types of dyes or pigments used in the mixture. Some markers generate a series of colored bands with known mobilities, while others produce a single band of a specific color that can be used as a reference point. They are widely used in research, clinical diagnostics, and forensic science.[4]
Progress markers
Loading buffers often contain anionic dyes that are visible under the visible light spectrum, and are added to the gel before the nucleic acid. Tracking dyes should not be reactive so as not to alter the sample, and move down the gel with the DNA or RNA sample.[5] Commonly used color markers include Bromophenol blue, Cresol Red, Orange G and Xylene cyanol. Xylene and bromophenol blue are the most commonly used dyes.[citation needed] Generally speaking, Orange G migrates faster than bromophenol blue, which migrates faster than xylene cyanol, but the apparent "sizes" of these dyes (compared to DNA molecules) varies with the concentration of agarose and the buffer system used. For instance, in a 1% agarose gel made in TAE buffer (Tris-acetate-EDTA), xylene cyanol migrates at the speed of a 3000 base pair (bp) molecule of DNA and bromophenol blue migrates at 400 bp. However, in a 1% gel made in TBE buffer (Tris-borate-EDTA), they migrate at 2000 bp and 250 bp respectively.[3]
DNA and RNA staining
Agarose gel electrophoresis is a technique widely used to estimate the size of nucleic acid fragments and identify them based on their differential mobility in the gel. Nucleic acids are commonly stained and detected using either ethidium bromide or SYBR Green dyes.[citation needed] The most common electrophoretic stain in agarose gel is ethidium bromide, however, SYBR green presents greater resolution and yield for single-stranded nucleic acid detection.[citation needed] The dyes grant fluorescence to DNA and RNA under 300 nm UV light. This occurs due to their intercalating nature. In double helical nucleic acids, the dyes bind between two strands, and in single-stranded nucleic acids, the dyes bind short, duplex segments formed within a strand.[citation needed]

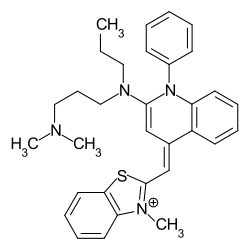
The most commonly used dye in agarose gel gel electrophoresis of DNA and RNA, dating as far back as the 1970s, is ethidium bromide (2,7-diamino-10-ethyl-9-phenylphenanthridiniumbromide).[citation needed] Ethidium Bromide (EtBr) is an orange-colored fluorescent intercalating dye. The dye inserts itself between the double helical structure of nucleic acids, allowing for visualization of the molecules under UV light.[6] EtBr has absorbance maxima at 300-360 nm and fluorescent emission maxima at 500-590 nm, with the detection limit of 0.5-5.0 ng/band.[6] The dye, however, has reduced sensitivity in the detection of single-stranded nucleic acid samples EtBr should be handled with care, as it is a potent mutagen.[6]
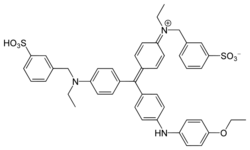
A more sensitive alternative for nucleic acid staining in gel electrophoresis is SYBR™ Green I.[7] The dye is 25 times more sensitive than EtBr in the staining of dsDNA, and is especially useful in staining assays containing single-stranded nucleic acids.[citation needed] SYBR Green is, however, more expensive when compared to EtBr.
Protein staining
Coomassie Blue is the most commonly used non-covalent stain in SDS polyacrylamide gel electrophoresis for protein quantification. The staining dye binds to the protein bands and creates a blue color that can be detected visually. Coomassie Brilliant Blue R-250 (red), is typically used for electrophoresis, while Coomassie Brilliant Blue G-250 (green), for Bradford Assay.[8] The limitation of this dye is that it is non-specific, and will bind to almost any protein in solution, and is less sensitive.[citation needed] Another common method of visualization of proteins in the gel is silver staining, where soluble silver ions permanently mark proteins and are reduced by formaldehyde to form a brown precipitate.[citation needed] Silver staining is a more sensitive staining method when compared to Coomassie Blue, however, results are more vulnerable to contamination.
Applications
Color markers are sometimes added to loading dyes for gel electrophoresis in the separation of DNA fragments. Loading dyes keep DNA samples below the surface of the agarose gel, and the color markers within help keep track of the migration front of the DNA as it moves along the gel.[9]
For PAGE, some commercially available molecular weight markers (also called "ladders" because they look like the rungs of a ladder after separation) contain pre-stained proteins of different colours, so it is possible to determine more accurately where the proteins of interest in the samples might be.
References
- ↑ "Products: Color Markers for Electrophoresis". Science 277 (5328): 979. 1997-08-15. doi:10.1126/science.277.5328.979. ISSN 0036-8075.
- ↑ 2.0 2.1 "Proposal of a laboratory course dedicated to the generation of protein molecular weight standards for sodium dodecyl sulfate-polyacrylamide gel electrophoresis". Biochemistry and Molecular Biology Education 49 (3): 353–360. May 2021. doi:10.1002/bmb.21476. PMID 33301651.
- ↑ 3.0 3.1 "A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans". Genetics 179 (1): 705–710. May 2008. doi:10.1534/genetics.108.087080. PMID 18493083.
- ↑ "Two-dimensional gel electrophoresis, isoelectric focusing and agarose gel electrophoresis in the diagnosis of multiple sclerosis". Journal of Clinical Chemistry and Clinical Biochemistry. Zeitschrift Fur Klinische Chemie und Klinische Biochemie 24 (12): 1017–1021. December 1986. doi:10.1515/cclm.1986.24.12.1017. PMID 3819655. http://edoc.hu-berlin.de/18452/11574.
- ↑ "Gel Electrophoresis Lab Procedures" (in en). 6 August 2018. https://sciencing.com/gel-electrophoresis-lab-procedures-5202505.html.
- ↑ 6.0 6.1 6.2 "Ethidium Bromide - US" (in en). https://www.thermofisher.com/us/en/home/life-science/dna-rna-purification-analysis/nucleic-acid-gel-electrophoresis/dna-stains/etbr.html.
- ↑ "SYBR™ Green I Nucleic Acid Gel Stain - 10,000X concentrate in DMSO". https://www.thermofisher.com/order/catalog/product/S7563.
- ↑ "Standard Dyes for Total Protein Staining in Gel-Based Proteomic Analysis". Materials 3 (10): 4784–4792. October 2010. doi:10.3390/ma3104784. PMID 28883353. Bibcode: 2010Mate....3.4784C.
- ↑ "Agarose gel electrophoresis for the separation of DNA fragments". Journal of Visualized Experiments 62 (62). April 2012. doi:10.3791/3923. PMID 22546956.
 |

