Biology:GIR1 branching ribozyme
The Lariat capping ribozyme (formerly called GIR1 branching ribozyme) is a ~180 nt ribozyme with an apparent resemblance to a group I ribozyme.[1] It is found within a complex type of group I introns also termed twin-ribozyme introns.[2] Rather than splicing, it catalyses a branching reaction in which the 2'OH of an internal residue is involved in a nucleophilic attack at a nearby phosphodiester bond.[3] As a result, the RNA is cleaved at an internal processing site (IPS), leaving a 3'OH and a downstream product with a 3 nt lariat at its 5' end. The lariat has the first and the third nucleotide joined by a 2',5' phosphodiester bond and is referred to as 'the lariat cap' because it caps an intron-encoded mRNA. The resulting lariat cap seems to contribute by increasing the half-life of the HE mRNA,[3][4] thus conferring an evolutionary advantage to the HE.
Biological context
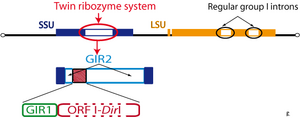
The GIR1 ribozyme was originally discovered during the functional characterization of the introns from the extrachromosomal rDNA of the Didymium iridis protist. A combination of deletion and in vitro self-splicing analyses revealed a twin-ribozyme intron organization: two distinct ribozyme domains within the intron.[2]
Structural organization
The twin-ribozyme introns represent some of the most complex organized group I introns known and consist of a homing endonuclease gene (HEG: I-DirI homing endonuclease) embedded in two functionally distinct catalytic RNA domains. One of the catalytic RNAs is a conventional group I intron ribozyme (GIR2) responsible for the intron splicing and reverse splicing, as well as intron RNA circularization. The other catalytic RNA domain is the group I-like ribozyme (GIR1) directly involved in homing endonuclease mRNA maturation.
Catalytic activity
| GIR1 Branching Ribozyme | |
|---|---|
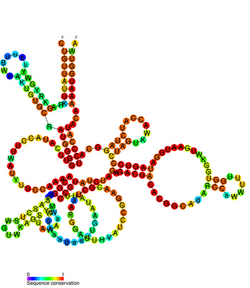 Conserved secondary structure of GIR1 | |
| Identifiers | |
| Symbol | GIR1 |
| Rfam | RF01807 |
| Other data | |
| RNA type | Intron |
| Domain(s) | Naegleria |
| PDB structures | PDBe |
In vitro, DiGIR1 catalyses three different reactions. The first one consists in hydrolysis of the scissile phosphate at the IPS site. This is the cleavage reaction observed with the full-length intron and several length variants with a relative low rate. The hydrolytic cleavage is irreversible and is considered an in vitro artefact resulting from misfolding of the catalytic site to present the branch nucleotide (BP) correctly for the reaction. The second reaction, the natural one, is the branching reaction, in which a transesterification at the IPS site results in the cleavage of the RNA with a 3'OH and a downstream lariat cap made by joining of the first and the third nucleotide by a 2'-5' phosphodiester bond.[3]
These products are the only products observed by analysis of cellular RNA.[4][5] This branching reaction is in equilibrium with a third one: a ligation reaction. It is a very efficient reaction and it tends to mask the branching reaction during the in vitro branching experiments with the full-length intron and length variants that include more than 166 nucleotides upstream of the IPS.
Modelling structure of the Lariat capping (LC) Ribozyme
GIR1 models have been created using biochemical and mutational data.[6] The structure contains an extended substrate domain which contains a GoU pair. The pair differs from the typical group 1 ribozyme nucleophilic residue, the J8/7 region has been reduced.[6] These findings provide the basis for an evolutionary mechanism that accounts for the change from group I splicing ribozyme to the branching GIR1 architecture. This mechanism could potentially be applied to other large RNAs such as the ribonuclease P.[6]
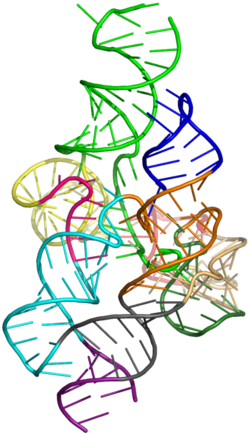
Crystal structure of the Lariat Capping Ribozyme
The crystal structure of the LC ribozyme was recently published.[7] In brief, a circularly permutated (CP) ribozyme RNA was generated by in vitro transcription using T7 RNA polymerase.[8] The 5' and 3' generated by circular permutation are located at the natural ribozyme cleavage site. To allow transcription of this construct, optimized 5' hammerhead and HdV (Hepatitis delay Virus) ribozymes were flanked to the LC CP construct.[9]
The crystal structure of the LC ribozyme unravels how the regulatory domain formed by P2, P2.1 and P10 works. Two sets of tertiary interactions take place to constrain P2 and P2.1 allowing the formation of a 3-way junction, which acts as a receptor for nt A209. This snug fit interaction promotes formation of the catalytic site, provided that the lariat is pre-folded by the ribozyme core.
References
- ↑ "DiGIR1 and NaGIR1: naturally occurring group I-like ribozymes with unique core organization and evolved biological role". Biochimie 84 (9): 905–12. September 2002. doi:10.1016/S0300-9084(02)01443-8. PMID 12458083.
- ↑ 2.0 2.1 "An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperate in self-splicing". Cell 76 (4): 725–34. February 1994. doi:10.1016/0092-8674(94)90511-8. PMID 8124711.
- ↑ 3.0 3.1 3.2 "An mRNA is capped by a 2', 5' lariat catalyzed by a group I-like ribozyme". Science 309 (5740): 1584–7. September 2005. doi:10.1126/science.1113645. PMID 16141078. Bibcode: 2005Sci...309.1584N.
- ↑ 4.0 4.1 "The group I-like ribozyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis". European Journal of Biochemistry 269 (23): 5804–12. December 2002. doi:10.1046/j.1432-1033.2002.03283.x. PMID 12444968.
- ↑ "In vivo expression of the nucleolar group I intron-encoded I-dirI homing endonuclease involves the removal of a spliceosomal intron". The EMBO Journal 18 (4): 1003–13. February 1999. doi:10.1093/emboj/18.4.1003. PMID 10022842.
- ↑ 6.0 6.1 6.2 "Molecular modelling of the GIR1 branching ribozyme gives new insight into evolution of structurally related ribozymes". The EMBO Journal 27 (4): 667–78. February 2008. doi:10.1038/emboj.2008.4. PMID 18219270.
- ↑ Meyer, M.; Nielsen, H.; Olieric, V.; Roblin, P.; Johansen, S. D.; Westhof, E.; Masquida, B. (2014-05-27). "Speciation of a group I intron into a lariat capping ribozyme". Proceedings of the National Academy of Sciences 111 (21): 7659–7664. doi:10.1073/pnas.1322248111. ISSN 0027-8424. PMID 24821772. Bibcode: 2014PNAS..111.7659M.
- ↑ Beckert, Bertrand; Masquida, Benoît (2011), Nielsen, Henrik, ed., "Synthesis of RNA by In Vitro Transcription", RNA: Methods and Protocols, Methods in Molecular Biology (Humana Press) 703: pp. 29–41, doi:10.1007/978-1-59745-248-9_3, ISBN 978-1-59745-248-9, PMID 21125481
- ↑ Meyer, Mélanie; Masquida, Benoît (2014), Waldsich, Christina, ed., "Cis-Acting 5' Hammerhead Ribozyme Optimization for In Vitro Transcription of Highly Structured RNAs", RNA Folding: Methods and Protocols, Methods in Molecular Biology (Humana Press) 1086: pp. 21–40, doi:10.1007/978-1-62703-667-2_2, ISBN 978-1-62703-667-2, PMID 24136596
Further reading
- "Book Review: Ribozymes and RNA Catalysis. Edited by David M. J. Lilley and Fritz Eckstein.". Angewandte Chemie International Edition 47 (45): 8558–9. 2008. doi:10.1002/anie.200885598.
- "Ribozyme structures and mechanisms". Annual Review of Biophysics and Biomolecular Structure 30: 457–75. 2001. doi:10.1146/annurev.biophys.30.1.457. PMID 11441810.
- "Evolution of biocatalysis 1. Possible pre-genetic-code RNA catalysts which are their own replicase". Origins of Life 14 (1–4): 291–300. 1984. doi:10.1007/BF00933670. PMID 6205343. Bibcode: 1984OrLi...14..291V. RNA catalysis evolutionary insight
External links
- Labs working on GIR1 branching ribozyme characterisation:
- Henrik Nielsen lab [1]
- Eric Westhof lab [2]
- Steinar Johansen lab [3]
- RNA catalysis
- Page for GIR1 branching ribozyme at Rfam
 |