Biology:Nodal signaling pathway
This article is missing information about nodal flow and its discovery [PMID 27821522]. (September 2021) |
The Nodal signaling pathway is a signal transduction pathway important in regional and cellular differentiation during embryonic development.[1]
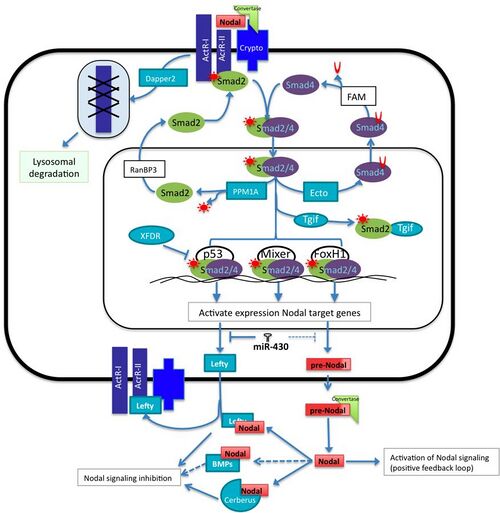
The Nodal family of proteins, a subset of the transforming growth factor beta (TGFβ) superfamily, is responsible for mesoendoderm induction, patterning of the nervous system, and determination of dorsal- ventral axis in vertebrate embryos. Activation of the Nodal pathway involves nodal binding to activin and activin-like receptors which leads to phosphorylation of the Smad2. The P-Smad2/Smad4 complex translocates into the nucleus to interact with transcription factors such as FoxH1, p53 and Mixer (Xenopus mix-like endodermal regulator). This will, in turn, lead to induction of target genes such as NODAL, Lefty, the antagonist of nodal cerberus, and others.[2]
The activation of the Nodal pathway induces the transcription of many target genes including of its own, but at the same time, micro-RNAs and other proteins interfere with this positive feedback loop in a negative manner at different points of the pathway.[2][3] This balance of activation and inhibition of the signal is necessary to achieve the precise location, concentration and duration of downstream target genes that have an important role early in development. This article will summarize the role of some of the components that participate positively and negatively in regulation the signaling pathway. Although all the major components of Nodal signaling are evolutionarily conserved in almost all vertebrates, the regulation of each component of the pathway sometimes varies according to the species.
History
The nodal gene was originally discovered by Conlon et al. by retroviral mutation in mice which led to the isolation of a gene that interfered with normal mouse gastrulation and embryo development.[4] Further study of this gene by Zhou et al. showed that the nodal genes encode a secreted signaling peptide that was sufficient to induce mesoderm cells in the mouse embryo. This was an important finding as many other factors had been implicated in the formation of mesoderm in Xenopus whereas the difficulty of removal of these factors due to embryonic lethality and maternal contribution of genes had kept the ability to assay the knock out phenotypes elusive.[5] Further studies of nodal signaling in other vertebrates such as Cyclops and Squint in zebrafish proved that nodal signaling is adequate to induce mesoderm in all vertebrates.[2]
Selected components of the pathway
Lefty
The Lefty proteins, divergent members of the TGFβ superfamily of proteins, act as extracellular antagonists of Nodal signaling. Expression studies of the Lefty homologue, antivin, in zebrafish show that Lefty likely acts as a competitive inhibitor of Nodal signaling.[6] Overexpression of Lefty leads to a phenotype similar to a Nodal knockout while overexpression of the activin (nodal-related protein) receptor or even the receptor extracellular domain can rescue the phenotype. As the induction of Lefty is dependent upon Nodal expression, lefty acts a classic feedback inhibitor for Nodal signaling. Like nodals, all vertebrates have at least one Lefty gene while many, such as zebrafish and mouse, have two unique Lefty genes.
DAN proteins
DAN proteins, such as Cerberus and Coco in Xenopus and Cerberus-like in mouse, also act as antagonists of Nodal signaling. Unlike Lefty proteins, DAN proteins bind directly to extracellular Nodal proteins and prevent signaling. Further, not all DAN proteins are specific to Nodal signaling and will also block bone morphogenetic proteins (BMPs) and, in the case of Cerberus and Coco, Wnt signaling as well.[7] This activity is important in neural development and left-right symmetry as will be discussed later.
BMPs
Lefty and Cerberus are not the only ones to be able to interact in the extracellular space with Nodal, there is biochemical evidence that BMP3 and BMP7 form heterodimers with Nodal, causing mutual inhibition of the involved pathways.[8]
Convertases: Furin and PACE4
Nodal mRNA produces an immature protein form of Nodal that is cleaved by proteins called convertases in order to generate a mature Nodal. The subtilisin-like proprotein convertases (SPC) Furin (Spc1) and PACE4 (Spc4) recognize a specific sequence of the precursor of Nodal protein and cleaves it to form the mature Nodal ligand.[9] Conversely, the immature form of Nodal is still capable to activate the pathway.[10] During Nodal transportation to the extracellular space, the Nodal co-receptor captures the Nodal precursor in lipid rafts and once in the cell surface, Cripto interacts with the convertases and forms a complex that facilitates the processing of Nodal.[11]
EGF-CFC proteins
EGF-CFC proteins are membrane bound extracellular factors that serve as essential cofactor in Nodal signaling and in vertebrate development as a whole. This family of cofactors includes One-eyed Pinhead (oep) in Zebrafish, FRL1 in Xenopus, and Cripto and Criptic in mouse and human. Genetic studies of oep in zebrafish have shown that the knockout of both maternal and zygotic oep leads to a phenotype similar to that of the squint/Cyclops (Nodals) knockout. Similarly, over-expression of either the Nodal (squint/Cyclops) or oep with the knockout of the other does not show phenotypical differences. This evidence coupled with the data that overexpression of oep shows no phenotype corroborates the role of EGF-CFC as an essential cofactor in Nodal signaling.[12]
Dapper2
In mouse, frog and fish, Dapper2, is a negative regulator of mesoderm formation acting through the down-regulation of the Wnt and TGFβ / nodal signaling pathways. In zebrafish, nodal is known to activate the gene expression of dapper2.[13] In the cell surface Dapper2 tightly binds to the active form of the activin type 1 receptors and targets the receptor for lysosomal degradation. Dapper2 overexpression mimics nodal co-receptor loss of function because nodal signal cannot be transduced and therefore it produces less mesoderm. In the mouse embryo, dpr2 mRNA is located across all the embryo 7.5 days post conception (dpc) however its location changes at 8.5-dpc where it is observed at the prospective somites and by 10-dpc, neural tube, otic vesicle and gut; because Dapper2 and Nodal are expressed in the same region, this suggests that Dapper antagonizes mesoderm induction signals derived from Nodal.[14] Somehow the reduction of activin receptors would lead to the decrease in activity of different TGFb pathways.[13]
Smad
Smad proteins are responsible for transducing nodal signals into the nucleus. The binding of Nodal proteins to activin or activin-like serine/threonine kinase receptors results in the phosphorylation of Smad2. Smad2 will then associate with Smad4 and translocate into the nucleus thereby stimulating transcription of nodal target genes. Evidence has been shown that another Smad, Smad3, can be phosphorylated by activated receptors and may also function as an activator of nodal genes. However, knockout of Smad2 in mice leads to disruption of the formation of the primitive streak. This is not sufficient to knockdown all mesoendodermal genes showing that Smad3 has some overlapping function with Smad2. However, the expression of these genes is ubiquitous in Smad2 KO embryos whereas it is limited in the wild type. Smad3 knockouts do not have a phenotype showing that expression overlap with Smad2 is sufficient normal development.[15]
Molecules affecting nodal activation via smad
Ectodermin negatively regulates the nodal pathway by inhibiting the interaction of Smad4 with other Smads inside the nucleus via the mono-ubiquitination Smad4, this modification allow it to be transported out of the cytoplasm where it can be deubiquitinated by FAM protein, allowing it to form complexes again with other Smads.[16][17] Another negative regulator of the pathway intervening with Smads is PPM1A, a phosphatase that acts with Phospho-Smad2/3 making it inactive.[18] Subsequently, Smad2/3 is transported outside the nucleus with the help of RanBP2.[19]
Transcriptional factors controlling signaling
Smad2/3/4 can associate to different transcription factors such as p53, Mixer and FoxH1 and recognize specific cis-regulatory elements to activate the expression of Nodal target genes at a precise time and location and activate genes required for mesoderm induction. There are some other transcription factors that compete for some of the components of the transcriptional machinery for the activation of Nodal target genes. For instance, Tgif1 and Tgif2 are negative co-regulators that compete for the active form of Smad2, reducing the relative concentration of active Smad2 in the nucleus. In Xenopus, the loss-of-function of Tgf1 and Tgf2 causes the up-regulation of Xnr5 and Xnr6.[20] Another example of transcriptional repressors in frog is XFDL, that binds to p53 obstructing the interaction with the Smad2/3/4 complex.[21]
miRNAs controlling signaling
In vertebrates, the evolutionary conserved family of microRNAs miR-430/427/302 is expressed early in development. It has important roles in controlling mesoderm and endoderm specification, and it does it by regulating the protein expression levels of some Nodal signaling components. This family is composed by the teleost miR-430, the amphibian miR-427 and the mammalian miR-302. In zebra fish the miR-430 inhibits translation of Sqt, Lefty1 and Lefty2, in frogs miR-427 regulate Xnr5, Xnr6b, LeftyA and LeftyB, however in humans embryonic stem cells it has been shown that miR-302 negative regulates the expression of only Lefty1 and Lefty2 but it does not seem down-regulate Nodal protein expression levels.[22]
Nodal signaling in development
Mesoendoderm Induction
Multiple studies have established that Nodal signal is required for the induction of most mesodermal and endodermal cell types and Squint/Cyclops knockouts in Zebrafish do not develop notochord, heart, kidneys or even blood.[23] The origin and expression pattern of the nodal signaling proteins differs in different species. Mammalian nodal signaling is initiated ubiquitously in epiblast cells and is maintained by autoregulatory signaling of Wnt3 and limited by the induction of antagonists such as Cerberus-like and lefty.[24] Studies in Xenopus have found that xnr expression (the Xenopus nodal) is induced by VegT at the vegetal pole and nodals spread to the blastula.[25] Xnr expression is stabilized by the presence of β-catenin. This information raises the question of how nodal signaling leads to the induction of both endoderm and mesoderm. The answer comes in form of a gradient of nodal protein. Temporal and spatial differences in nodal signaling will result in different cell fates. With the addition of antagonists and variable range of different nodals, a map of cell fates including both mesoderm and endoderm can be drawn for the embryo.[2] However, it is unclear whether nodal signaling is summated or if cells respond to the amplitude of the signal.[26]
Left-Right Patterning
Human anatomy is asymmetric with the heart located on the left side and the liver on the right. Left-right asymmetry is a feature common to all vertebrates and even paired-symmetric organs such as lungs display asymmetries in the number of lobes. Evidence that nodal signaling is responsible for left-right specification comes from genetic analysis of organisms deficient in left-right specification. These genetic studies led to identification of mutations in components in the nodal signaling pathway such as ActRIIB, Criptic, and FoxH1 in mouse.[27] These studies found that the left-right symmetry is created as a result of nodal antagonist expression on the right side of the embryo which is balanced by nodal upregulating itself on the other half of the embryo. The result is a nodal gradient that is high on the ventral side of the embryo and, through antagonist action, declines as a gradient to the midline. Studies on the nodal signaling pathway and its downstream targets such as PITX2 in other animals have shown it may also control left-right asymmetric patterning in sea squirt, amphioxus, sea urchin and mollusc lineages. [28]
Neural patterning
As nodal signaling give rise to ectoderm and mesoderm, neuroectoderm formation requires blocking nodal signaling which is accomplished by the expression of nodal antagonist, Cerberus. The role of nodal signaling reemerges later in development when nodal signaling is required to specify ventral cell neural patterning. Loss of function of Cyclops or oep in zebrafish results in cyclopic embryos characterized by a lack of medial floor plate and ventral forebrain.[2] Not all nodals result in the formation of mesoectoderm. Xenopus nodal related 3, (Xnr3) a divergent member of the TGFβ superfamily, induces the expression of the protein, Xbra. The Xbra expression pattern, in correlation the expression pattern another neuroinducer, Xlim-1, result in the patterning of the organizer in Xenopus. This signaling in conjuncture with other nodals, noggin, chordin, follistatin and others results in the final patterning of vertebrate central nervous system.[29]
References
- ↑ Shen MM (March 2007). "Nodal signaling: developmental roles and regulation". Development 134 (6): 1023–34. doi:10.1242/dev.000166. PMID 17287255.
- ↑ 2.0 2.1 2.2 2.3 2.4 Schier AF (2003). "Nodal signaling in vertebrate development". Annu. Rev. Cell Dev. Biol. 19: 589–621. doi:10.1146/annurev.cellbio.19.041603.094522. PMID 14570583.
- ↑ Schier AF (November 2009). "Nodal morphogens". Cold Spring Harb Perspect Biol 1 (5): a003459. doi:10.1101/cshperspect.a003459. PMID 20066122.
- ↑ "A novel retrovirally induced embryonic lethal mutation in the mouse: assessment of the developmental fate of embryonic stem cells homozygous for the 413.d proviral integration". Development 111 (4): 969–81. April 1991. doi:10.1242/dev.111.4.969. PMID 1879365.
- ↑ "Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation". Nature 361 (6412): 543–7. February 1993. doi:10.1038/361543a0. PMID 8429908. Bibcode: 1993Natur.361..543Z. https://zenodo.org/record/1233139.
- ↑ "Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction". Development 126 (2): 229–40. January 1999. doi:10.1242/dev.126.2.229. PMID 9847237.
- ↑ "The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals". Nature 397 (6721): 707–10. February 1999. doi:10.1038/17820. PMID 10067895. Bibcode: 1999Natur.397..707P.
- ↑ "Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms". Mol. Cell 7 (5): 949–57. May 2001. doi:10.1016/S1097-2765(01)00249-0. PMID 11389842.
- ↑ "Extraembryonic proteases regulate Nodal signalling during gastrulation". Nat. Cell Biol. 4 (12): 981–5. December 2002. doi:10.1038/ncb890. PMID 12447384.
- ↑ "The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4". Dev. Cell 11 (3): 313–23. September 2006. doi:10.1016/j.devcel.2006.07.005. PMID 16950123.
- ↑ "Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation". EMBO J. 27 (19): 2580–91. October 2008. doi:10.1038/emboj.2008.174. PMID 18772886.
- ↑ "The EGF-CFC gene family in vertebrate development". Trends Genet. 16 (7): 303–9. July 2000. doi:10.1016/S0168-9525(00)02006-0. PMID 10858660.
- ↑ 13.0 13.1 Chen YG (January 2009). "Endocytic regulation of TGF-beta signaling". Cell Res. 19 (1): 58–70. doi:10.1038/cr.2008.315. PMID 19050695.
- ↑ "The evolutionally conserved activity of Dapper2 in antagonizing TGF-beta signaling". FASEB J. 21 (3): 682–90. March 2007. doi:10.1096/fj.06-6246com. PMID 17197390.
- ↑ Whitman M (November 2001). "Nodal signaling in early vertebrate embryos: themes and variations". Dev. Cell 1 (5): 605–17. doi:10.1016/S1534-5807(01)00076-4. PMID 11709181.
- ↑ "Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase". Cell 121 (1): 87–99. April 2005. doi:10.1016/j.cell.2005.01.033. PMID 15820681.
- ↑ "FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination". Cell 136 (1): 123–35. January 2009. doi:10.1016/j.cell.2008.10.051. PMID 19135894.
- ↑ "PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling". Cell 125 (5): 915–28. June 2006. doi:10.1016/j.cell.2006.03.044. PMID 16751101.
- ↑ "Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling". Dev. Cell 16 (3): 345–57. March 2009. doi:10.1016/j.devcel.2009.01.022. PMID 19289081.
- ↑ "Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation". Development 137 (2): 249–59. January 2010. doi:10.1242/dev.040782. PMID 20040491.
- ↑ "Ectodermal factor restricts mesoderm differentiation by inhibiting p53". Cell 133 (5): 878–90. May 2008. doi:10.1016/j.cell.2008.03.035. PMID 18510931.
- ↑ "The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection". Dev. Cell 16 (4): 517–27. April 2009. doi:10.1016/j.devcel.2009.02.007. PMID 19386261.
- ↑ "Nodal signaling patterns the organizer". Development 127 (5): 921–32. March 2000. doi:10.1242/dev.127.5.921. PMID 10662632.
- ↑ "Nodal signalling in the epiblast patterns the early mouse embryo". Nature 411 (6840): 965–9. June 2001. doi:10.1038/35082103. PMID 11418863. Bibcode: 2001Natur.411..965B.
- ↑ "Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors". Development 126 (24): 5759–70. December 1999. doi:10.1242/dev.126.24.5759. PMID 10572051.
- ↑ Green J (December 2002). "Morphogen gradients, positional information, and Xenopus: interplay of theory and experiment". Dev. Dyn. 225 (4): 392–408. doi:10.1002/dvdy.10170. PMID 12454918.
- ↑ "Conserved and divergent mechanisms in left-right axis formation". Genes Dev. 14 (7): 763–76. April 2000. doi:10.1101/gad.14.7.763. PMID 10766733.
- ↑ Namigai, E; Kenny NJ; Shimeld SM (2014). "Right across the tree of life: the evolution of left–right asymmetry in the Bilateria". Genesis 52 (6): 458–470. doi:10.1002/dvg.22748. PMID 24510729.
- ↑ "Role of the Xlim-1 and Xbra genes in anteroposterior patterning of neural tissue by the head and trunk organizer". Proc. Natl. Acad. Sci. U.S.A. 94 (3): 895–900. February 1997. doi:10.1073/pnas.94.3.895. PMID 9023353. Bibcode: 1997PNAS...94..895T.
Further reading
- "Direct and indirect roles for Nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart". Proc. Natl. Acad. Sci. U.S.A. 105 (37): 13924–9. September 2008. doi:10.1073/pnas.0802159105. PMID 18784369. Bibcode: 2008PNAS..10513924B.
- "Nodal signaling and the evolution of deuterostome gastrulation". Dev. Dyn. 234 (2): 269–78. October 2005. doi:10.1002/dvdy.20549. PMID 16127715.
- "The zebrafish Nodal signal Squint functions as a morphogen". Nature 411 (6837): 607–10. May 2001. doi:10.1038/35079121. PMID 11385578. Bibcode: 2001Natur.411..607C.
- "The role of the zebrafish nodal-related genes squint and cyclops in patterning of mesendoderm". Development 130 (9): 1837–51. May 2003. doi:10.1242/dev.00400. PMID 12642489.
- "Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway". Development 137 (3): 417–26. February 2010. doi:10.1242/dev.039735. PMID 20056679.
 |

