Biology:OCC-1
OCC-1 (overexpressed in colon carcinoma-1) is a protein, which in humans is encoded by the gene C12orf75.[1] The gene is approximately 40,882 bp long and encodes 63 amino acids. OCC-1 is ubiquitously expressed throughout the human body. OCC-1 has shown to be overexpressed in various colon carcinomas.[2] Novel splice variant of this gene was also detected in various human cancer types; in addition to encoding a novel smaller protein (51 amino acids), OCC-1 gene produces a non-protein coding RNA splice variant lncRNA (called OCC-D variant).
Gene
Location and size: C12orf75 is found along the plus strand of chromosome 12 (12q23.3).[3] The gene is 40,882 bp long with the genomic sequence beginning at 105,330,636 bp and ends at 105,371,518 bp. C12orf75 contains 6 exons and is flanked by KCCAT198 (renal clear cell carcinoma-associated transcript 198).[4]
mRNA
C12orf75 encodes several transcripts of mRNA that the longest one is 1386 bp.[5] The mature mRNA of this splice variant contains six exons.
Protein
- Structure
Primary structure: OCC-1 protein is 63 amino acids long [6] and has a molecular weight of 6.4 kdal.[7] OCC-1 contains an “opioid growth factor receptor repeat” (OGFr) [8] motif from residue 8 to 27. OCC-1 is an acidic protein with an isoelectric point of OCC-1 is 6.6.[9]
Secondary structure: The secondary structure of OCC-1 is a combination of multiple coils, a few α-helices, and few β-sheets. The Phyre 2 program 52% α-helices, 6% β-sheets, and 70% disordered.[10] The predicted region β-sheet from residues 31 to 38 coincide with the α-helices and β-sheets regions predicted by other programs. OCC-1 is a soluble protein; according to the SOUSI program therefore there are no transmembrane domains.[11]
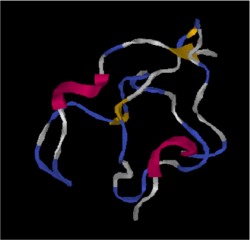
Tertiary structure: Predicted folding by iTASSER is shown.[12]
Post translational modifications: OCC-1 is predicted to undergo the post-translational modifications of O-glycosylation, phosphorylation, and myristoylation.[13]
| Type of Modification | Amino Acid Position | Impact on Protein |
|---|---|---|
| O-N-acetyleglucosamine | Ser6, Thr9, Ser10, Thr25, Ser54 | Nucleocytoplasmic location, cell signaling |
| Phosphorylation | Ser28, Tyr42, Thr59 | Conformational change, enzymatic functions of protein |
| Myristoylation | Gly2, Gly4, Gly40, Gly44 | Allows for weak protein-protein and protein-lipid interactions |
Subcellular location: The k-NN tool places OCC-1 in the nucleus of the cell with 65.2% certainty, 13% in the mitochondria, 8.7% in the vesicles of the secretory system, 4.3% cytoplasm, and 4.3% vacuole.
Homology
Paralogs: OCC-1 has no known paralogs.
Orthologs: OCC-1 has been found in mammalia, reptilia, amphibian, aves, and actinopterygii.[14] The gene is not found in plants, protists, fungi, archaea, or bacteria. The most distant ortholog is the Oryzias latipers or the Japanese rice fish, which diverged from the human gene approximately 436.5 million years ago.[15]
Phylogeny: The phylogenetic tree to the right shows the evolution of OCC-1 among humans and the orthologs from the various taxa that contain OCC-1. The results of this phylogenetic tree follow in accordance with the predicted evolutionary history of animals on Earth.
Expression
Expression level: OCC-1 has moderate to high expression throughout the body, therefore OCC-1 is ubiquitously expressed in humans; notably high expression in the kidney, skeletal muscle and pancreas and low expression in the heart. OCC-1 has shown to be overexpressed in various colon carcinomas.[16] In regards to homologous expression, in situ hybridization data revealed that OCC-1 is expressed in the primary visual cortex of the macque, in an activity dependent manner.[17]
Disease state expression: Profiles from NCBI UniGene show the expression of OCC-1 in adrenal tumors, chondrosarcoma, gastrointestinal tumors, kidney tumors, leukemia, liver tumors, prostate cancer, soft tissue/muscle tissue tumors, and uterine tumors.[18]
Regulation of expression
Promoter: The promoter of OCC-1 is GXP_4407929 and 601 bp in length.[19] The promoter can be found on the plus strand and begins at 105234790 bp and ends at 105235390 bp.
Interacting proteins
OCC-1 is shown to interact with HRG4, ELAVL1, c-REL, and IRS4. ELAVL1 functions to stabilized mRNA for gene expression. C-REL is involved in lymphoid and cell growth/survival, with a specific presence in T cell malignancies and cancer.[20] HRG4 plays a role in signal transduction and trafficking sensory neurons and is located primarily in the retina,[21] which relates to the discovery of expression in the brain of the macque through in situ hybridization data. IRS4 functions as interface between growth factor receptors consisting of tyrosine kinase activity, such as insulin receptors.[22] IRS4 also is involved with the IGF1R mitogenic signaling pathway.
Clinical significance
OCC-1 has shown to be overexpressed in multiple colon carcinomas.[citation needed]
References
- ↑ "C12orf75 chromosome 12 open reading frame 75 [Homo sapiens (human)] – Gene – NCBI". https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=387882. Retrieved 2016-05-05.
- ↑ Pibouin, Laurence; Villaudy, Jacqueline; Ferbus, Didier; Muleris, Martine; Prospéri, Marie-Thérèse; Remvikos, Yurgos; Goubin, Gérard (2002). "Cloning of the mRNA of overexpression in colon carcinoma-1". Cancer Genetics and Cytogenetics 133 (1): 55–60. doi:10.1016/S0165-4608(01)00634-3. PMID 11890990.
- ↑ "C12orf75 Gene (Protein Coding) : Chromosome 12 Open Reading Frame 75". https://www.genecards.org/cgi-bin/carddisp.pl?gene=C12orf75. Retrieved 2016-05-05.
- ↑ "KCCAT198 renal clear cell carcinoma-associated transcript 198 [Homo sapiens (human)] – Gene – NCBI". https://www.ncbi.nlm.nih.gov/gene/105369954. Retrieved 2016-05-05.
- ↑ "Homo sapiens chromosome 12 open reading frame 75 (C12orf75), mRNA – Nucleotide – NCBI". 2015-09-28. https://www.ncbi.nlm.nih.gov/nuccore/NM_001145199.1. Retrieved 2016-05-05.
- ↑ "overexpressed in colon carcinoma 1 protein [Homo sapiens] – Protein – NCBI". 2015-09-28. https://www.ncbi.nlm.nih.gov/protein/223633952?report=fasta. Retrieved 2016-05-05.
- ↑ "SDSC Biology Workbench". http://workbench.sdsc.edu/. Retrieved 2016-05-05.
- ↑ "Motif Scan". http://myhits.isb-sib.ch/cgi-bin/motif_scan. Retrieved 2016-05-05.
- ↑ "SDSC Biology Workbench". Workbench.sdsc.edu. http://workbench.sdsc.edu/. Retrieved 2016-05-05.
- ↑ Lawrence Kelley (2011-02-14). "PHYRE2 Protein Fold Recognition Server". http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index. Retrieved 2016-05-05.
- ↑ [1][|permanent dead link|dead link}}]
- ↑ "I-TASSER server for protein structure and function prediction". http://zhanglab.ccmb.med.umich.edu/I-TASSER/. Retrieved 2016-05-05.
- ↑ "SIB Bioinformatics Resource Portal – Resources". ExPASy. 2015-07-31. http://www.expasy.org/resources/search/querytext:secondary%20structure. Retrieved 2016-05-05.
- ↑ "BLAST: Basic Local Alignment Search Tool". 2016-01-20. http://blast.ncbi.nlm.nih.gov/Blast.cgi. Retrieved 2016-05-05.
- ↑ "The Timescale of Life". TimeTree. http://www.timetree.org/. Retrieved 2016-05-05.
- ↑ "Home – Geo – Ncbi". https://www.ncbi.nlm.nih.gov/geo/. Retrieved 2016-05-05.
- ↑ Tochitani, Shiro; Liang, Fengyi; Watakabe, Akiya; Hashikawa, Tsutomu; Yamamori, Tetsuo (2001). "Theocc1gene is preferentially expressed in the primary visual cortex in an activity-dependent manner: A pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex". European Journal of Neuroscience 13 (2): 297–307. doi:10.1046/j.0953-816x.2000.01390.x. PMID 11168534.
- ↑ "Home – UniGene – NCBI". 2015-09-28. https://www.ncbi.nlm.nih.gov/unigene. Retrieved 2016-05-05.
- ↑ "Genomatix: Login Page". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=dea7e029d7c3843bc63f162e099d45de;SHOW_ANNOTATION=C12orf75;ELDORADO_VERSION=E31R1508. Retrieved 2016-05-05.
- ↑ Gilmore, T. D.; Gerondakis, S. (2011). "The c-Rel Transcription Factor in Development and Disease". Genes & Cancer 2 (7): 695–711. doi:10.1177/1947601911421925. PMID 22207895.
- ↑ Higashide, Tomomi; Inana, George (1999). "Characterization of the Gene for HRG4 (UNC119), a Novel Photoreceptor Synaptic Protein Homologous to Unc-119". Genomics 57 (3): 446–50. doi:10.1006/geno.1999.5791. PMID 10329014.
- ↑ "IRS4 – Insulin receptor substrate 4 – Homo sapiens (Human) – IRS4 gene & protein". https://www.uniprot.org/uniprot/O14654. Retrieved 2016-05-05.
 |