Biology:Origin of transfer
An origin of transfer (oriT) is a short sequence ranging from 40-500 base pairs in length[1][2] that is necessary for the transfer of DNA from a gram-negative bacterial donor to recipient during bacterial conjugation.[3][4][5] The transfer of DNA is a critical component for antimicrobial resistance within bacterial cells[6] and the oriT structure and mechanism within plasmid DNA is complementary to its function in bacterial conjugation. The first oriT to be identified and cloned was on the RK2 (IncP) conjugative plasmid, which was done by Guiney and Helinski in 1979.[7]
Structure
oriT regions are central to the process of transferring DNA from the donor to recipient and contain several important regions that facilitate this:
- nic site: where the unwound plasmid DNA is cut; usually site-specific.[4][8][9]
- An inverted repeat sequence: signals the end of replication of donor DNA and is responsible for transfer frequency, plasmid mobilization, and secondary DNA structure formation.[3][8][10]
- AT-rich region: important for DNA strand opening and is located adjacent to the inverted repeat sequences.[1][3][5][8][11][12]
The oriT is a noncoding region of the bacterial DNA.[13] Due to its important role in initiating bacterial conjugation, the oriT is both an enzymatic substrate and recognition site for the relaxase proteins.[1][13][14] Relaxosomes have oriT-specific auxiliary factors that help it to identify and bind to the oriT.[1] Upstream of the oriT nic site is a termination sequence.[5]
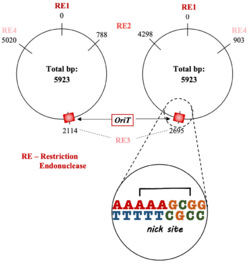
oriTs are primarily cis-acting, which allows for a more efficient DNA transfer.[5][12][15]

Mechanism and function in bacterial conjugation
At the start of bacterial conjugation, a donor cell will elaborate a pilus and signal to a nearby recipient cell to get in close contact. This identification of a suitable recipient cell will begin the mating pair formation process.[1][16] This process of bringing the two cells together recruits the type IV secretion system, a protein complex that forms the transfer channel between the donor and recipient, starting the formation of the relaxation complex known as the relaxosome at the oriT.[13]
A plasmid's oriT sequence serves as both a recognition point and a substrate for the enzymes in the relaxosome,[13] therefore the first step of bacterial conjugation occurs at the nicn site of the oriT region of the plasmid.[4][14] Relaxase enzymes, otherwise known as DNA strand transferases part of the relaxosome complex, catalyze a strand- and site-specific phosphodiester bond cleavage at the nicn site and are specific to each plasmid.[17] This reaction is a trans-esterification, which produces a nicked double-stranded DNA with the 5' end bound to a tyrosine residue in the relaxase.[4][5][14][17] The relaxase then moves toward the 3' end of the strand to unwind the DNA in the plasmid.[17]
The other strand of the plasmid, the strand that was not nicked by the relaxase, is a template for further synthesis by DNA polymerase.[17]
Once the relaxase reaches the upstream section of the oriT again where there is an inverted repeat, the process is terminated by reuniting the ends of the plasmid and releasing a single-stranded plasmid in the recipient.[5][15][18]
Applications
Genetic engineering
Conjugation allows for the transfer of target genes to many recipients, including yeast,[19] mammalian cells,[20][21] and diatoms.[22]
Diatoms could be useful plasmid hosts as they have the potential to autotrophically produce biofuels and other chemicals.[22] There are some methods for genetic transfer for diatoms, but they are slow compared to bacterial conjugation. By designing plasmids for the diatoms P. tricornutum and T. pseudonana based on sequences for yeast and developing a method for conjugation from E. coli to the diatoms, researchers hope to advance genetic manipulation in diatoms.[22]
One of the main problems in using bacterial conjugation in genetic engineering is that certain selectable markers on the plasmids generate bacteria that have resistance to antibiotics like ampicillin and kanamycin.[23]
Antimicrobial resistance
The interaction between the DNA oriT and relaxase enables antimicrobial resistance via horizontal gene transfer (Figure 1).[13] Various oriT regions in plasmid DNA contain inverted repeats onto which relaxase proteins are able bind.[3] Major contributors of drug resistance are mobile genomic islands (MGIs), or segments in DNA that are found in similar strains of bacteria and are factors in diversification of bacteria.[3][24] MGIs provide resistance to their host cells, and through bacterial conjugation, spread this advantage to other cells.[3] With bacterial cell MGIs having their own oriT sequences and being in close proximity to relaxosome genes, they are very similar to conjugative plasmids that are responsible for the prevalence of drug resistance among bacterial cells.[3] A 2017 study on MGIs revealed that they are able to integrate themselves into the genome of the receiving bacterial cells by themselves via int, a gene that codes for the integrase enzyme. After the oriT of the MGI are processed by the relaxosomes encoded by integrative and conjugative elements (ICE), the MGI are able to enter the genome of the receiver cells and allow for the multiformity of bacteria that leads to antimicrobial resistance.[24]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "Conjugative DNA metabolism in Gram-negative bacteria". FEMS Microbiology Reviews 34 (1): 18–40. January 2010. doi:10.1111/j.1574-6976.2009.00195.x. PMID 19919603.
- ↑ Frost, L. S. (2009-01-01), Schaechter, Moselio, ed. (in en), Conjugation, Bacterial, Oxford: Academic Press, pp. 517–531, doi:10.1016/b978-012373944-5.00007-9, ISBN 978-0-12-373944-5, https://www.sciencedirect.com/science/article/pii/B9780123739445000079, retrieved 2021-12-03
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "Identification and Characterization of oriT and Two Mobilization Genes Required for Conjugative Transfer of Salmonella Genomic Island 1". Frontiers in Microbiology 10: 457. 2019. doi:10.3389/fmicb.2019.00457. PMID 30894848.
- ↑ 4.0 4.1 4.2 4.3 "Stepwise assembly of a relaxosome at the F plasmid origin of transfer" (in English). The Journal of Biological Chemistry 270 (47): 28381–28386. November 1995. doi:10.1074/jbc.270.47.28381. PMID 7499340.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 "DNA processing reactions in bacterial conjugation". Annual Review of Biochemistry 64 (1): 141–169. June 1995. doi:10.1146/annurev.bi.64.070195.001041. PMID 7574478.
- ↑ "Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease". Veterinary Pathology 51 (2): 328–340. March 2014. doi:10.1177/0300985813511131. PMID 24318976.
- ↑ "The DNA-protein relaxation complex of the plasmid RK2: location of the site-specific nick in the region of the proposed origin of transfer". Molecular & General Genetics 176 (2): 183–189. October 1979. doi:10.1007/BF00273212. PMID 393953.
- ↑ 8.0 8.1 8.2 "A classification scheme for mobilization regions of bacterial plasmids". FEMS Microbiology Reviews 28 (1): 79–100. February 2004. doi:10.1016/j.femsre.2003.09.001. PMID 14975531.
- ↑ "The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer". Molecular Microbiology 25 (3): 509–516. August 1997. doi:10.1046/j.1365-2958.1997.4861849.x. PMID 9302013.
- ↑ "In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins". Nucleic Acids Research 20 (1): 41–48. January 1992. doi:10.1093/nar/20.1.41. PMID 1738602.
- ↑ "The origin of transfer (oriT) of the conjugative plasmid R46: characterization by deletion analysis and DNA sequencing". Molecular & General Genetics 208 (1–2): 219–225. June 1987. doi:10.1007/BF00330445. PMID 3039307.
- ↑ 12.0 12.1 "Deletion analysis of the F plasmid oriT locus". Journal of Bacteriology 173 (3): 1012–1020. February 1991. doi:10.1128/jb.173.3.1012-1020.1991. PMID 1991706.
- ↑ 13.0 13.1 13.2 13.3 13.4 "DNA structure at the plasmid origin-of-transfer indicates its potential transfer range". Scientific Reports 8 (1): 1820. January 2018. doi:10.1038/s41598-018-20157-y. PMID 29379098. Bibcode: 2018NatSR...8.1820Z.
- ↑ 14.0 14.1 14.2 "Nicking by transesterification: the reaction catalysed by a relaxase". Molecular Microbiology 25 (6): 1011–1022. September 1997. doi:10.1046/j.1365-2958.1997.5241885.x. PMID 9350859.
- ↑ 15.0 15.1 "Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis". Journal of Bacteriology 189 (20): 7254–7261. October 2007. doi:10.1128/JB.00932-07. PMID 17693500.
- ↑ "F conjugation: back to the beginning". Plasmid. Special Issue based on the International Society for Plasmid Biology Meeting: Santander 2012 70 (1): 18–32. July 2013. doi:10.1016/j.plasmid.2013.03.010. PMID 23632276.
- ↑ 17.0 17.1 17.2 17.3 "Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC". Nature Structural Biology 10 (12): 1002–1010. December 2003. doi:10.1038/nsb1017. PMID 14625590.
- ↑ "Analysis of the sequence and gene products of the transfer region of the F sex factor". Microbiological Reviews 58 (2): 162–210. June 1994. doi:10.1128/mr.58.2.162-210.1994. PMID 7915817.
- ↑ "Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast". Nature 340 (6230): 205–209. July 1989. doi:10.1038/340205a0. PMID 2666856. Bibcode: 1989Natur.340..205H.
- ↑ "Genetic transformation of HeLa cells by Agrobacterium". Proceedings of the National Academy of Sciences of the United States of America 98 (4): 1871–1876. February 2001. doi:10.1073/pnas.98.4.1871. PMID 11172043. Bibcode: 2001PNAS...98.1871K.
- ↑ "Conjugation between bacterial and mammalian cells". Nature Genetics 29 (4): 375–376. December 2001. doi:10.1038/ng779. PMID 11726922.
- ↑ 22.0 22.1 22.2 "Designer diatom episomes delivered by bacterial conjugation". Nature Communications 6 (1): 6925. April 2015. doi:10.1038/ncomms7925. PMID 25897682. Bibcode: 2015NatCo...6.6925K.
- ↑ "Persistence and reversal of plasmid-mediated antibiotic resistance". Nature Communications 8 (1): 1689. November 2017. doi:10.1038/s41467-017-01532-1. PMID 29162798. Bibcode: 2017NatCo...8.1689L.
- ↑ 24.0 24.1 "Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits". Mobile Genetic Elements 7 (2): 1–6. 2017-03-04. doi:10.1080/2159256X.2017.1304193. PMID 28439449.
 |
