Biology:Personalized genomics
Personalized genomics is the human genetics-derived study of analyzing and interpreting individualized genetic information by genome sequencing to identify genetic variations compared to the library of known sequences. International genetics communities have spared no effort from the past and have gradually cooperated to prosecute research projects to determine DNA sequences of the human genome using DNA sequencing techniques. The methods that are the most commonly used are whole exome sequencing and whole genome sequencing. Both approaches are used to identify genetic variations. Genome sequencing became more cost-effective over time, and made it applicable in the medical field, allowing scientists to understand which genes are attributed to specific diseases.
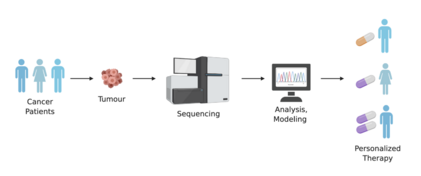
Personalized medicine is an emerging practice in medicine that develops patient-specific treatments based on an individual's genetic profile. The treatment enables patients to experience maximized therapeutic effectiveness and minimized adverse effects. Personalized medicine has been widely accepted, and future-oriented changes in policy and infrastructure are implemented throughout the world to readily adopt into other fields.
History
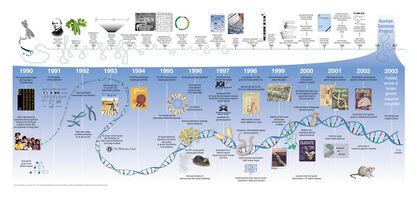
Efforts to explore genes and heredity have been prolonged for over 100 years. From Gregor Mendel’s studies of inheritance,[1] many researchers have dedicated themselves to scientific development via new discoveries and inventions, such as the DNA double helix discovered by Rosalind Franklin and the sanger sequencing[2] invented by Frederick Sanger. Recognition of chromosomal disorders like trisomy in 1959-1960[3] has led scientists to realize that genes are related to the phenotypic distinctive diseases. It motivated scientists all over to carry out several projects together to understand the human genome.
Human Genome Project
Human Genome Project (HGP)[4] is a research project conducted by universities and research centers throughout six countries with the primary goal of determining the complete sequence of bases of the entire human genome and identifying the complete set of human genes. This project also stored the genetic information in public databases and had a role in the improvement of tools for data analysis. A group of anonymous human volunteers recruited from diverse populations was sequenced. The establishment of HGP accelerated tracing human origins by detecting genetic mutations and constructing genetic maps. Additionally, it helped to launch other research projects around the world.
International HapMap Project
International HapMap Project[5] is an international research project facilitating identification of genotype-phenotype correlation by genome-wide association study (GWAS). This research created a haplotype map of the human genome to find genetic variations that affect human disease from 11 global populations. The project focused mainly on single nucleotide polymorphisms (SNPs) distributed within and across populations, and designed high density SNP arrays to screen for polymorphisms, allowing researchers to search for genes involved in common human diseases.
These projects have been completed with the aid of DNA sequencing techniques which allow for the identification of genetic variations in each genome, thus personally diagnosing genetic diseases.
Genome analysis

Personalized genomics includes the analysis of the genome of individuals.[2] To analyze the genome, a basic knowledge of genetics is required. There are DNA building blocks that are called nucleotides and those nucleotides are annealed to form a strand. Two strands of DNA are supercoiled into double helix form and that is what people think of first when they imagine the DNA structure. These DNA are very important as they contain genetic information. The order of nucleotides determines the genomics of individuals, and individuals have different genomics .[2] Therefore, people have different characteristics such as eye color. Not only characteristics can be revealed by DNA, but different causes of unique diseases can be identified by determining the order of nucleotides through DNA sequencing.
Whole exome and genome sequencing
To analyze personal genomics, a technique called DNA sequencing is needed and it is used to determine any disorders or polymorphisms in DNA sequences. There are two methods to conduct DNA sequencing, Whole Exome Sequencing (WES)[2] and Whole Genome Sequencing (WGS).[6] Formal way of sequencing, the sanger technique had some limitations that it was costly and time-consuming. The recent development of Next Generation Sequencing (NGS)[7] dramatically remedied the shortcomings of Sanger sequencing.
NGS enabled the sequencing of large loads of DNA.[7] There are pieces of genes that transcribe the proteins. These pieces of gene give instructions to make complementary proteins and are called exons. Exons are known to make up 1 percent of a human's genome.[2] All the exons in a genome together are called the exome. Whole exome sequencing is the method to sequence all exons in genomics. As proteins produced by the exons are crucial for human metabolism and the endosomal environment, if mutations occur at the exons, it will lead to a critical genetic disease. Hence, most mutations that cause known genetic diseases arise in the exon area. WES is an efficient way to sequence possible disease-causing mutations.
WGS, on the other hand, literally stands for the method that sequences every nucleotide of an individual's DNA and can detect any variations in any location in the genome.[6] Researchers have found that mutations at outside of exon areas also can influence gene transcription and protein production.[8] In other words, DNA variation at the non-exon area can induce genetic disorders that WES can not detect. Hence, there are studies that conduct both WES and WGS together to increase the quality of genomic data to identify genetic disorders.[9]
By WES and WGS, various genetic variations can be observed compared to select-gene sequencing, but the significance of the information obtained from whole-scale sequencing is mostly unknown. This is because there is no 100% causal link between genetic variation and health.[2] There are some mutations that do not affect human health or cause disease. It cannot be deduced that identified genetic change is an inducing factor of the disease of interest. Hence, a large sample size is needed to be analyzed to identify causal variants. However, the usage of next-generation sequencing has revolutionized the individualized healthcare regimens used to treat rare diseases using personalized medicine.
Applications
Genetic medicine
Genetic medicine refers to the application of genetics to medical care in the detection and treatment of several phenotypic rare hereditary disorders, and this discipline incorporates an emerging area in medicine such as personalized medicine.
Personalized medicine
Personalized medicine is a type of tailored medical treatment to the individual patients’ characteristics based on their expected response or resistance to diseases. While traditional medicine follows a ‘one-size-fits-all’ approach[10] that drug is designed to treat a large population of patients, personalized medicine is prescribed to the group of patients organized by molecular diagnostic and analytic techniques relating to their genetic information to minimize adverse effects and to maximize treatment effectiveness.
Necessity
The necessity for personalized medicine becomes particularly evident with the reduction in the therapeutic effectiveness of existing medicine. In a BBC news interview with Allen D. Roses, a former senior vice president at GlaxoSmithKline[11] claimed that 90% of drugs have a therapeutic effect on only 30-50% of patients. Patients have experienced varying success rates from drugs distributed in the market with a low efficacy rate of 50% or below.[11] He emphasized that different genetic information contributes to variability in the response to the medication between patients. As an example of cancer cells, tumor heterogeneity has been found to be associated with the clinical outcome of cancer immunotherapy,[12] so diagnostic techniques with high accuracy must be utilized.
Biomarker
Accurate diagnosis of patients’ circumstances must be performed to discover biomarkers, which are biological indicators of medical state which can be measured objectively and accurately. A prognostic signature developed by biomarker discovery is predictive of disease progression and patients’ response to drug intervention. Consequently, it can be used in the development of new pharmaceuticals and in the selection of eligible patients who are expected to be treated with certain medications. For example, single molecular array technology[13] has detected changes in prostate-specific antigen levels between each prostate cancer cell. Moreover, fiber microarrays[13] have been served as a multiplexed diagnostics platform for identifying potential biomarkers of cystic fibrosis, including interleukin-8,[14] vascular endothelial growth factor,[15] epidermal growth factor,[13] interferon-gamma inducible protein,[13] and matrix metallopeptidase-9.[13] These biomarker signatures enabled patients to be differentiated into subgroups depending on their disease severity.
Examples
The integration of diagnostic techniques and genomic sequencing has played a vital role in allowing for the implementation of personalized medicine in the market. For instance, warfarin[16] may lead to acute cardiac failure, hemorrhage, necrosis, and osteoporosis depending on the genetic variations in the CYP2C9,[16] and VKORC1 gene.[16] Since the prescription dose for warfarin varies by approximately 10-fold based on the genetic variation of each patient, U.S. Food and Drug Administration (FDA)[16] recommends that warfarin dosing should be prescribed after conducting genetic testing. In addition, gefitinib, an inhibitor of epidermal growth factor receptor (EGFR) -tyrosine kinase, has therapeutic efficacy on only breast cancer[17] or non-small cell lung cancer.[18] Genetic variations in the EGFR gene have been reported to be associated with the sensitivity of patients with lung cancer to gefitinib.[19] Patients with breast cancer expressing activated HER2/neu have benefited from therapeutic effects of gefitinib.[20] FDA-approved[21] gefitinib should be given for the treatment of patients with breast or non-small cell lung cancer.
Future trends
Cost of the sequencing
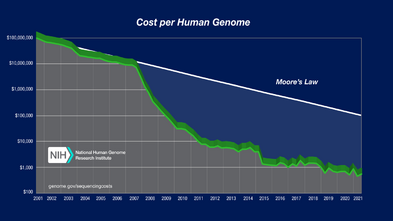
The price of DNA sequencing was very expensive until recently. WGS cost about $20,000 during the 2010s.[22] WGS and WES are relatively recent technologies in clinical practice. Current data of health economic evidence to encourage use of WES and WGS in clinical research is restricted. Hence, there are not enough studies that carefully evaluate and indicate the cost-effectiveness of these technologies and components that are included in cost estimates. However, the development of Artificial Intelligence (AI) and the knowledge of big data and machine learning,[13] enabled researchers to analyze massive loads of DNA sequencing faster and efficiently. Now the price of WGS fell to its lowest, $1,500. Single tests for WES range from $555 to $5,169 and for WGS from $1,906 to $24,810.[23] It is decreasing with the development of technologies. Currently, due to the notable decrease in the cost, countries apart from the developed countries, are planning to do WGS of the unique patients or by conducting joint study to get a foundation to form a big data of population genetics.
Shift in infrastructure
The importance of engineering and computation has been brought to the attention of clinics and the marketplace in compliance with their incorporation into the medical field and growing public demand for individualized health care services. Computer-based analytical platforms using AI,[13] and Machine Learning[13] are practicable for clinical trial use. They were able to determine patient-specific optimal therapies based on subpopulation-specific stratifications classified by genome sequencing. According to the study guided by the AI platform,[24] liver transplant immunosuppressant prescriptions were successfully recommended depending on the patient's genetic information, and it confirmed that there was less inter-patients variance in AI-derived drug recommendation compared to control cohort patients. Computational analytical platforms predicted patients’ response towards drugs and boosted the process of recreating medical drugs and device validation.[13] The integration of computer programming and personalized medicine enables a large number of patients to receive personal care in a short period of time with greater efficiency.
Political Change
The medical community follows an increasing trend of translating traditional medicine into personalized medicine, and long-held regulatory policies are obstacles to be resolved for its public usage. The United States took action via the enactment of the 21st Century Cures Act in 2016.[13] This enforcement provided financial aid to U.S. Food and Drug Administration (FDA),[13] allowing to accelerate the emergence of newly personalized products such as cell therapy and medical devices into the marketplace. Also, National Institutes of Health (NIH) benefited from the resource allocation. Many countries have introduced legislation for the improvement of personalized medicine. In Singapore, AI Singapore, a national AI initiative supported by National Research Foundation,[13] was able to enhance national AI capabilities in drug development and patient-centered care. In addition, the Innovative Medicines Initiative (IMI)[13] was established between the European Union and pharmaceutical companies with the purpose of developing medical devices, drugs, and vaccines to tackle major challenges that will imperil the EU.
See also
- Precision medicine
- Genetic privacy
- Population genetics
- Reference genome
- Pharmacogenomics
- Personalized onco-genomics
References
- ↑ "Mendel as the Father of Genetics :: DNA from the Beginning". http://www.dnaftb.org/1/bio.html.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 "What are whole exome sequencing and whole genome sequencing?: MedlinePlus Genetics" (in en). https://medlineplus.gov/genetics/understanding/testing/sequencing/.
- ↑ "Origins of human genetics. A personal perspective". European Journal of Human Genetics 29 (7): 1038–1044. July 2021. doi:10.1038/s41431-020-00785-7. PMID 33542497.
- ↑ "A Journey Through The History Of DNA Sequencing" (in en-US). 2020-11-02. https://the-dna-universe.com/2020/11/02/a-journey-through-the-history-of-dna-sequencing/.
- ↑ International HapMap Consortium (December 2003). "The International HapMap Project". Nature 426 (6968): 789–796. doi:10.1038/nature02168. PMID 14685227.
- ↑ 6.0 6.1 "Whole Genome Sequencing" (in en). Encyclopedia of Gerontology and Population Aging. Cham: Springer International Publishing. 2021. pp. 5424–5433. doi:10.1007/978-3-030-22009-9_943. ISBN 978-3-030-22008-2. https://link.springer.com/10.1007/978-3-030-22009-9_943. Retrieved 2022-03-29.
- ↑ 7.0 7.1 "Genome-Wide Sequencing for Unexplained Developmental Disabilities or Multiple Congenital Anomalies: A Health Technology Assessment". Ontario Health Technology Assessment Series 20 (11): 1–178. 2020. PMID 32194879.
- ↑ "Whole genome sequencing analysis for cancer genomics and precision medicine". Cancer Science 109 (3): 513–522. March 2018. doi:10.1111/cas.13505. PMID 29345757.
- ↑ "The genetic structure of the Turkish population reveals high levels of variation and admixture". Proceedings of the National Academy of Sciences of the United States of America 118 (36): e2026076118. September 2021. doi:10.1073/pnas.2026076118. PMID 34426522. Bibcode: 2021PNAS..11826076K.
- ↑ "One Size Does Not Fit All: A Comprehensive Clinical Approach to Reducing Suicidal Ideation, Attempts, and Deaths". International Journal of Environmental Research and Public Health 16 (19): 3606. September 2019. doi:10.3390/ijerph16193606. PMID 31561488.
- ↑ 11.0 11.1 "Drugs 'don't work on many people'" (in en-GB). 2003-12-08. http://news.bbc.co.uk/2/hi/health/3299945.stm.
- ↑ "Intratumoral heterogeneity in cancer progression and response to immunotherapy". Nature Medicine 27 (2): 212–224. February 2021. doi:10.1038/s41591-021-01233-9. PMID 33574607.
- ↑ 13.00 13.01 13.02 13.03 13.04 13.05 13.06 13.07 13.08 13.09 13.10 13.11 13.12 "Enabling Technologies for Personalized and Precision Medicine". Trends in Biotechnology 38 (5): 497–518. May 2020. doi:10.1016/j.tibtech.2019.12.021. PMID 31980301.
- ↑ "Correlations of salivary biomarkers with clinical assessments in patients with cystic fibrosis". PLOS ONE 10 (8): e0135237. 2015. doi:10.1371/journal.pone.0135237. PMID 26258476. Bibcode: 2015PLoSO..1035237N.
- ↑ "Multiplexed salivary protein profiling for patients with respiratory diseases using fiber-optic bundles and fluorescent antibody-based microarrays". Analytical Chemistry 85 (19): 9272–9280. October 2013. doi:10.1021/ac4019523. PMID 23972398.
- ↑ 16.0 16.1 16.2 16.3 "Warfarin Therapy and VKORC1 and CYP Genotype". Medical Genetics Summaries [Internet].. Bethesda (MD): National Center for Biotechnology Information (US). 2012. http://www.ncbi.nlm.nih.gov/books/NBK84174/. Retrieved 2022-04-20.
- ↑ "The efficacy of gefitinib supplementation for breast cancer: A meta-analysis of randomized controlled studies" (in en-US). Medicine 99 (43): e22613. October 2020. doi:10.1097/MD.0000000000022613. PMID 33120749.
- ↑ "Gefitinib (Iressa) | Cancer information | Cancer Research UK". https://www.cancerresearchuk.org/about-cancer/cancer-in-general/treatment/cancer-drugs/drugs/gefitinib#:~:text=Gefitinib%20is%20a%20treatment%20for,trials%20for%20other%20cancer%20types..
- ↑ "EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib". Proceedings of the National Academy of Sciences of the United States of America 101 (36): 13306–13311. September 2004. doi:10.1073/pnas.0405220101. PMID 15329413. Bibcode: 2004PNAS..10113306P.
- ↑ "Breast cancer expressing the activated HER2/neu is sensitive to gefitinib in vitro and in vivo and acquires resistance through a novel point mutation in the HER2/neu". Cancer Research 67 (14): 6825–6843. July 2007. doi:10.1158/0008-5472.CAN-07-0765. PMID 17638894.
- ↑ "FDA Approval of Gefitinib for the Treatment of Patients with Metastatic EGFR Mutation-Positive Non-Small Cell Lung Cancer". Clinical Cancer Research 22 (6): 1307–1312. March 2016. doi:10.1158/1078-0432.CCR-15-2266. PMID 26980062.
- ↑ "Personal Genome Test Will Sell at New Low Price of $250" (in en). https://www.scientificamerican.com/article/craig-venter-s-company-in-deal-for-whole-exome-tests-at-new-low-cost/.
- ↑ "Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature". Genetics in Medicine 20 (10): 1122–1130. October 2018. doi:10.1038/gim.2017.247. PMID 29446766.
- ↑ "CURATE.AI: Optimizing Personalized Medicine with Artificial Intelligence". SLAS Technology 25 (2): 95–105. April 2020. doi:10.1177/2472630319890316. PMID 31771394.
 |