Biology:Phasevarion
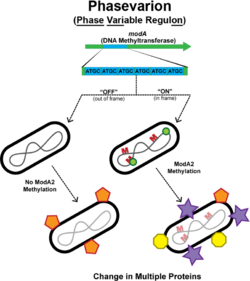
In bacteria, phasevarions (also known as phase variable regulons) mediate a coordinated change in the expression of multiple genes or proteins.[1] This occurs via phase variation of a single DNA methyltransferase. Phase variation of methyltransferase expression results in differential methylation throughout the bacterial genome, leading to variable expression of multiple genes through epigenetic mechanisms.
Phasevarions have been identified in several mucosal-associated human-adapted pathogens, which include; Haemophilus influenzae,[2] Neisseria meningitidis,[3] Neisseria gonorrhoeae,[3] Helicobacter pylori,[4] Moraxella catarrhalis,[5] and Streptococcus pneumoniae.[6] All described phasevarions regulate expression of proteins that are involved in host colonization, survival, and pathogenesis, and many regulate putative vaccine targets.[7] The presence of phasevarions complicates identification of stably expressed proteins, as the regulated genes do not contain any identifiable features. The only way to identify genes in a phasevarion is by detailed study of the organisms containing such systems. Study of the phasevarions, and identification of proteins they regulate, is therefore critical to generate effective and stable vaccines.
Phase variable DNA methyltransferases
Many of the phasevarions described to date are controlled by Type III methyltransferases.[8] Mod genes are the methyltransferase component of type III restriction modification (R-M) systems in bacteria, and serve to protect host DNA from the action of the associated restriction enzyme. However, in many bacterial pathogens, mod genes contain simple sequence repeats (SSRs), and the associated restriction enzyme encoding gene (res) is inactive. In these organisms the DNA methyltransferase phase varies between two states (ON or OFF) by variation in the number of SSRs in the mod gene.[9] Multiple different mod genes have been identified. Each Mod methylates a different DNA sequence in the genome. Methylation of unique DNA sequences results in different Mod enzymes that regulate the expression of different sets of genes; i.e., they control different phasevarions. For example, twenty-one unique modA alleles have been identified in Haemophilus influenzae;[10][11] Neisseria species contain seven modB alleles;[12] and Helicobacter pylori contains seventeen modH alleles.[4] Individual strains of Neisseria gonorrhoeae and Neisseria meningitidis can contain multiple, independently switching mod genes; for example, N. gonorrhoae can contain both modA and modB genes002C and individual N. meningitidis strains that contain modA, modB and modD have been identified.[12][13]
A phasevarion controlled by a methyltransferase associated with a Type I R-M system has been identified and studied in Streptococcus pneumoniae.[6] This phase-variable methyltransferase switches between six different methyltransferase specificities by shuffling between multiple, variable copies of the specificity subunit, hsdS, that dictates the sequence to be methylated. By shuffling DNA sequences, six different HsdS specificity proteins are produced in a pneumococcal population. This means six different DNA sequences are methylated by the functional methyltransferase. This genetic shuffling, or recombination, occurs between inverted repeat sequences located in the multiple, variable hsd genes present in the locus. Recombination is catalyzed by a recombinase that is associated with the type I locus. These six methyltransferase specificities (SpnD39IIIA-F) result in six differentiated cell types in a pneumococcal population.[14][6]
A potential phasevarion controlled by a Type IIG R-M system has been recently described in the human gastric pathogen Campylobacter jejuni.[15]
Role in pathogenesis
Switching of mod genes is selected for under certain disease states or within specific host niches: for example, the non-typeable Haemophilus influenzae (NTHi) modA2 ON state is selected for within the middle ear during manifestation of experimental otitis media.[11] A switch from modA2 OFF to modA2 ON results in more severe middle ear disease in a model of otitis media than in a situation where switching from modA2 OFF to modA2 ON does not occur.[16] Phase-variation of the modA2 allele also results in NTHi populations with distinct advantages under oxidative stress and increased resistance to neutrophil killing.[17] In M. catarrhalis, the modM3 allele is associated with strains isolated from the middle ear of children.[5] In S. pneumoniae, selection of particular SpnD39III alleles (allele A) occurs when S. pneumoniae is present in blood, which implies that SpnD39III-A regulates genes that give a selective advantage in this in vivo niche. No selection for any SpnD39III allele was seen when S. pneumoniae was present in the nasopharynx.[6]
References
- ↑ "The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes". Proc Natl Acad Sci U S A 102 (15): 5547–51. 2005. doi:10.1073/pnas.0501169102. PMID 15802471.
- ↑ "Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression". Nucleic Acids Res 35 (15): 5242–52. 2007. doi:10.1093/nar/gkm571. PMID 17675301.
- ↑ 3.0 3.1 "Phasevarions mediate random switching of gene expression in pathogenic Neisseria". PLOS Pathog 5 (4): e1000400. 2009. doi:10.1371/journal.ppat.1000400. PMID 19390608.
- ↑ 4.0 4.1 "Phasevarion mediated epigenetic gene regulation in Helicobacter pylori". PLOS ONE 6 (12): e27569. 2011. doi:10.1371/journal.pone.0027569. PMID 22162751.
- ↑ 5.0 5.1 "ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media". FASEB J. 28 (12): 5197–207. 2014. doi:10.1096/fj.14-256578. PMID 25183669. http://www.fasebj.org/content/28/12/5197.
- ↑ 6.0 6.1 6.2 6.3 "A random six-phase switch regulates pneumococcal virulence via global epigenetic changes". Nat. Commun. 5 (5): 5055. 2014. doi:10.1038/ncomms6055. PMID 25268848.
- ↑ "The Capricious Nature of Bacterial Pathogens: Phasevarions and Vaccine Development". Front. Immunol. 7 (7): 586. 2016. doi:10.3389/fimmu.2016.00586. PMID 28018352.
- ↑ "Type III restriction-modification enzymes: a historical perspective". Nucleic Acids Res. 42 (1): 45–55. 2014. doi:10.1093/nar/gkt616. PMID 23863841.
- ↑ "The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes". Nat Rev Microbiol 8 (3): 196–206. 2010. doi:10.1038/nrmicro2283. PMID 20140025.
- ↑ "Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae". PLOS ONE 7 (3): e32337. 2012. doi:10.1371/journal.pone.0032337. PMID 22457715.
- ↑ 11.0 11.1 "A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae". Nat. Commun. 6: 7828. 2015. doi:10.1038/ncomms8828. PMID 26215614.
- ↑ 12.0 12.1 "Distribution of the type III DNA methyltransferases modA, modB and modD among Neisseria meningitidis genotypes: implications for gene regulation and virulence". Sci. Rep. 6: 21015. 2016. doi:10.1038/srep21015. PMID 26867950.
- ↑ "Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N(6)-adenine DNA methyltransferases of Neisseria meningitidis". Nucleic Acids Res. 43 (8): 4150–62. 2015. doi:10.1093/nar/gkv219. PMID 25845594.
- ↑ "A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes". Nucleic Acids Res. 31 (7): 1805–12. 2003. doi:10.1093/nar/gkg274. PMID 12654995.
- ↑ Anjum, A; Brathwaite, KJ; Aidley, J; Connerton, PL; Cummings, NJ; Parkhill, J; Connerton, I; Bayliss, CD (2 June 2016). "Phase variation of a Type IIG restriction-modification enzyme alters site-specific methylation patterns and gene expression in Campylobacter jejuni strain NCTC11168.". Nucleic Acids Research 44 (10): 4581–94. doi:10.1093/nar/gkw019. PMID 26786317.
- ↑ "ModA2 Phasevarion Switching in Nontypeable Haemophilus influenzae Increases the Severity of Experimental Otitis Media". J Infect Dis 214 (5): 817–24. 2016. doi:10.1093/infdis/jiw243. PMID 27288538.
- ↑ "The ModA2 Phasevarion of nontypeable Haemophilus influenzae Regulates Resistance to Oxidative Stress and Killing by Human Neutrophils". Sci Rep 7 (1): 3161. 2017. doi:10.1038/s41598-017-03552-9. PMID 28600561.
 |
