Biology:Haemophilus influenzae
| Haemophilus influenzae | |
|---|---|
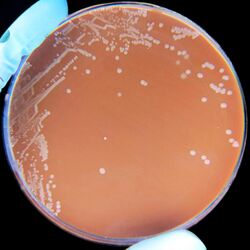
| |
| H. influenzae on a chocolate agar plate | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota |
| Class: | Gammaproteobacteria |
| Order: | Pasteurellales |
| Family: | Pasteurellaceae |
| Genus: | Haemophilus |
| Species: | H. influenzae
|
| Binomial name | |
| Haemophilus influenzae (Lehmann & Neumann 1896)
Winslow et al. 1917 | |
Haemophilus influenzae (formerly called Pfeiffer's bacillus or Bacillus influenzae) is a Gram-negative, non-motile, coccobacillary, facultatively anaerobic, capnophilic pathogenic bacterium of the family Pasteurellaceae. The bacteria are mesophilic and grow best at temperatures between 35 and 37 °C.[1]
H. influenzae was first described in 1893[2][3] by Richard Pfeiffer during an influenza pandemic[4] when he incorrectly identified it as the causative microbe, which is why the bacteria was given the name "influenzae".[5][6] H. influenzae is responsible for a wide range of localized and invasive infections, typically in infants and children,[7] including pneumonia, meningitis, or bloodstream infections.[8] Treatment consists of antibiotics; however, H. influenzae is often resistant to the penicillin family, but amoxicillin/clavulanic acid can be used in mild cases.[9] Serotype B H. influenzae have been a major cause of meningitis in infants and small children, frequently causing deafness and mental retardation. However the development in the 1980s of a vaccine effective in this age group (the Hib vaccine) has almost eliminated this in developed countries.
This species was the first organism to have its entire genome sequenced.[10][11]
Physiology and metabolism
Structure
H. influenzae is a small Gram-negative bacterium, approximately 0.3 micrometer to 1 micrometer.[12] Like other Gram-negative bacteria, H. influenzae has a thin peptidoglycan layer surrounded by an outer membrane containing lipopolysaccharide.[13] Some types of H. influenzae contain a polysaccharide capsule around the outer membrane to aid in protection and colonization.[14] The bacteria are pleomorphic, meaning the shape of the bacterium is variable, however it is typically coccobacillus or rod-shaped.[15] H. Influenzae contains pili, which are specialized to adhere to the human nasopharynx. The H. Influenzae pili, unlike those of E. coli, resist unwinding, allowing for stronger adhesion to resist expulsion when coughing or sneezing.[16] A minority of non-typeable, or unencapsulated, H. influenzae employ a variety of attachment techniques, such as pili, adhesins, or Hia and Hap proteins.[17] Though the bacteria possess pili, they are not used for traditional movement or motility, and the bacterium is still considered to be non-motile.[18]
The cell wall of H. influenzae bacterium contains various proteins, referred to as autotransporters, for adherence and colony formation. H. influenzae prefers to bind to mucus linings or non-ciliated epithelial cells, which is facilitated by Hap𝘴 autotransporters in the cell wall binding with unknown receptors within the epithelium.[19] The Hap𝘴 autotransporters also facilitate the formation of microcolonies of the bacteria. These microcolonies are likely responsible for the formation of various biofilms within the body, such as those responsible for middle ear or lung infections.[19]
Penicillin binding proteins
Penicillin binding proteins (PBPs) catalyze steps in peptidoglycan metabolism. They carry out essential processes needed to build and modify the cell wall.[20] These proteins are the targets blocked by penicillin and other beta-lactam antibiotics that bind to PBPs, hence their name.[21] Some antibiotic-resistant isolates of H. Influenzae contain modified PBPs that resist beta-lactam action by producing beta-lactamases to degrade these antibiotics. This resistance is likely due to a N526K mutation, or R517H substitution in conjunction with another unknown mutation. The R517H substitution alone did not have a lower affinity for penicillin, and therefore cannot cause resistance alone.[20] Beta-lactamase emergence in the 1970s caused the therapy for severe cases of H. influenzae to be changed from ampicillin to cephalosporins, however further resistance to cephalosporins has occurred due to changes in the transpeptidase domain of penicillin binding protein 3 (PBP3).[22]
Serotypes
H. influenzae isolates were initially characterized as either encapsulated (having an extracellular polysaccharide layer, the bacterial capsule) or unencapsulated. Encapsulated strains were further classified on the basis of the immune response to the type of polysaccharides in their capsule. The six generally recognized types of encapsulated H. influenzae are: a, b, c, d, e, and f.[23] H. Influenzae type b, also known as Hib, is the most common form, recognizable by its polyribosyl ribitol phosphate (PRP) capsule, and found mostly in children.[24] Types a, e, and f have been isolated infrequently, while types d and c are rarely isolated. Unencapsulated strains are more genetically diverse than the encapsulated group.[25] Unencapsulated strains are termed nontypable (NTHi) because they lack capsular serotypes; however, all H. influenzae isolates can now be classified by multilocus sequence typing and other molecular methods. Most NTHi strains are considered to be part of the normal human flora in the upper and lower respiratory tract, genitals, and conjunctivae (mucous membranes of the eye).[24]
Metabolism
H. influenzae uses the Embden–Meyerhof–Parnas (EMP) pathway for glycolysis and the pentose phosphate pathway, which is anabolic rather than catabolic. The citric acid cycle is incomplete and lacks several enzymes that are found in a fully functioning cycle. The enzymes missing from the TCA cycle are citrate synthase, aconitate hydratase, and isocitrate dehydrogenase.[26] H. influenzae has been found in both aerobic and anaerobic environments, as well as environments with different pH's.[27]
Genome and genetics
H. influenzae was the first free-living organism to have its entire genome sequenced. Completed by Craig Venter and his team at The Institute for Genomic Research, now part of the J. Craig Venter Institute. Haemophilus was chosen because one of the project leaders, Nobel laureate Hamilton Smith, had been working on it for decades and was able to provide high-quality DNA libraries. The sequencing method used was whole-genome shotgun, which was completed and published in Science in 1995.[10]
The genome of strain Rd KW20 consists of 1,830,138 base pairs of DNA in a single circular chromosome that contains 1604 protein-coding genes, 117 pseudogenes, 57 tRNA genes, and 23 other RNA genes.[10] About 90% of the genes have homologs in E. coli, another gamma-proteobacterium. In fact, the similarity between genes of the two species ranges from 18% to 98% protein sequence identity, with the majority sharing 40-80% of their amino acids (with an average of 59%).[28]
Conjugative plasmids (DNA molecules that are capable of horizontal transfer between different species of bacteria) can frequently be found in H. influenzae. It is common that the F+ plasmid of a competent Escherichia coli bacterium conjugates into the H. influenzae bacterium, which then allows the plasmid to transfer among H. influenzae strands via conjugation.[29]
Role of transformation
H. influenzae mutants defective in their rec1 gene (a homolog of recA) are very susceptible to being killed by the oxidizing agent hydrogen peroxide.[30] This finding suggests that rec1 expression is important for H. influenzae survival under conditions of oxidative stress. Since it is a homolog of recA, rec1 likely plays a key role in recombinational repair of DNA damage. Thus, H. influenzae may protect its genome against the reactive oxygen species produced by the host's phagocytic cells through recombinational repair of oxidative DNA damages.[31] Recombinational repair of a damaged site of a chromosome requires, in addition to rec1, a second homologous undamaged DNA molecule. Individual H. influenzae cells are capable of taking up homologous DNA from other cells by the process of transformation. Transformation in H. influenzae involves at least 15 gene products,[10] and is likely an adaptation for repairing DNA damage in the resident chromosome.[32]
Culture methods and diagnosis of infections
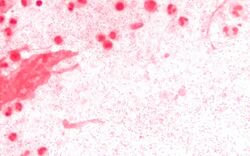

Culture
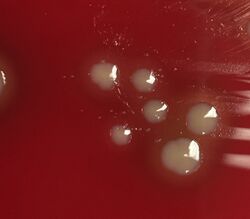
Bacterial culture of H. influenzae is performed on agar plates. The strongest growth is seen on chocolate agar at 37 °C in a CO2-enriched incubator.[33] The ideal CO2 concentration for the culture is ~5%.[34] However adequate growth is often seen on brain-heart infusion agar supplemented with , with added hemin and nicotinamide adenine dinucleotide (NAD)
Colonies of H. influenzae appear as convex, smooth, pale, grey, or transparent colonies with a mild odor.[34] H. influenzae will only grow on blood agar if other bacteria are present to release these factors from the red blood cells, forming 'satellite' colonies around these bacteria. For example, H. influenzae will grow in the hemolytic zone of Staphylococcus aureus on blood agar plates; the hemolysis of cells by S. aureus releases NAD which is needed for its growth. H. influenzae will not grow outside the hemolytic zone of S. aureus due to the lack of nutrients in these areas.[35]
Diagnosis of infections
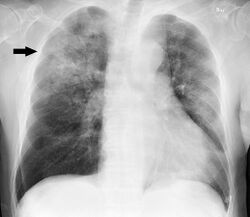
Clinical features of a respiratory tract infection may include initial symptoms of an upper respiratory tract infection mimicking a viral infection, usually associated with low-grade fevers. This may progress to the lower respiratory tract within a few days, with features often resembling those of wheezy bronchitis. Sputum may be difficult to expectorate and is often grey or creamy in color. The cough may persist for weeks without appropriate treatment. Many cases are diagnosed after presenting chest infections that do not respond to penicillins or first-generation cephalosporins. A chest X-ray can identify alveolar consolidation.[36]
Clinical diagnosis of invasive H. influenzae infection (infection that has spread to the bloodstream and internal tissues) is typically confirmed by bacterial culture, latex particle agglutination tests, or polymerase chain reaction tests on clinical samples obtained from an otherwise sterile body site. In this respect, H. influenzae cultured from the nasopharyngeal cavity or throat would not indicate H. influenzae disease, because these sites are colonized in disease-free individuals.[37] However, H. influenzae isolated from cerebrospinal fluid or blood or joint fluid would indicate invasive H. influenzae infection.Microscopic observation of a Gram stained specimen of H. influenzae will show Gram-negative coccobacillus. The cultured organism can be further characterized using catalase and oxidase tests, both of which should be positive. Further serological testing is necessary to distinguish the capsular polysaccharide and differentiate between H. influenzae b and nonencapsulated strains.[citation needed]
Although highly specific, bacterial culture of H. influenzae lacks sensitivity. Use of antibiotics prior to sample collection greatly reduces the isolation rate by killing the bacteria before identification is possible.[38] Recent work has shown that H. influenzae uses a highly specialized spectrum of nutrients where lactate is a preferred carbon source.[39]
Latex particle agglutination
The latex particle agglutination test (LAT) is a more sensitive method to detect H. influenzae than is culture.[40] Because the method relies on antigen rather than viable bacteria, the results are not disrupted by prior antibiotic use. It also has the added benefit of being quicker than culture methods. However, antibiotic sensitivity testing is not possible with LAT alone, so a parallel culture is necessary.[41]
Molecular methods
Polymerase chain reaction (PCR) assays have been proven to be more sensitive than either LAT or culture tests and are highly specific.[42] These PCR tests can be used for capsular typing of encapsulated H. influenzae strains.[43]
Pathogenicity
Host colonization
Many microbes colonize within a host organism. Colonization occurs when a microorganism continues to multiply within the host, without interaction, causing no visible signs of illness or infection. H. influenzae colonizes differently in adults than it does young children. Because this bacterium colonizes more rapidly in young children, they are capable of carrying more than one strain of the same bacterium. Once in the adult stage of life, a human is likely to only be carrying one strain as this bacterium does not colonize as aggressively in adults. Nearly all infants will undergo colonization of this bacteria within their first year of life.[17]
H. influenzae is generally found within and upon the human body, but can also live on various dry, hard surfaces for up to 12 days.[44][45] Most strains of H. influenzae are opportunistic pathogens; that is, they usually live in their host without causing disease, but cause problems only when other factors (such as a viral infection, reduced immune function or chronically inflamed tissues, e.g. from allergies) create an opportunity. They infect the host by sticking to the host cell using trimeric autotransporter adhesins.[46]
The pathogenesis of H. influenzae infections is not completely understood, although the presence of the polyribosyl ribitol phosphate (PRP) capsule in encapsulated type b (Hib), a serotype causing conditions such as epiglottitis, is known to be a major factor in virulence.[47] Their capsule allows them to resist phagocytosis and complement-mediated lysis in the nonimmune host. The unencapsulated strains are almost always less invasive; however, they can produce an inflammatory response in humans, which can lead to many symptoms. Vaccination with Hib conjugate vaccine is effective in preventing Hib infection but does not prevent infection with NTHi strains.[48]
H. influenzae can cause respiratory tract infections including pneumonia, otitis media, epiglottitis (swelling in the throat), eye infections and bloodstream infection, meningitis. It can also cause cellulitis (skin infection) and infectious arthritis (inflammation of the joint).[49]
Haemophilus influenzae type b (Hib) infection
Naturally acquired disease caused by H. influenzae seems to occur in humans only. In healthy children under the age of 5, H. influenzae type b was responsible for more than 80% of aggressive infections, before the introduction of the [Hib] vaccine.[50] In infants and young children, H. influenzae type b (Hib) causes bacteremia, pneumonia, epiglottitis and acute bacterial meningitis.[51] On occasion, it causes cellulitis, osteomyelitis, and infectious arthritis. It is one cause of neonatal infection.[52]
Due to routine use of the Hib vaccine in the U.S. since 1990, the incidence of invasive Hib disease has decreased to 1.3/100,000 in children.[51] However, Hib remains a major cause of lower respiratory tract infections in infants and children in developing countries where the vaccine is not widely used. Unencapsulated H. influenzae strains are unaffected by the Hib vaccine and cause ear infections (otitis media), eye infections (conjunctivitis), and sinusitis in children, and are associated with pneumonia.[51]
Treatment
Some strains of H. influenzae produce beta-lactamases, and are also able to modify its penicillin-binding proteins, so the bacteria have gained resistance to the penicillin family of antibiotics. In severe cases, cefotaxime and ceftriaxone delivered directly into the bloodstream are the elected antibiotics, and, for the less severe cases, an association of ampicillin and sulbactam, cephalosporins of the second and third generation, or fluoroquinolones are preferred. (Fluoroquinolone-resistant strains of H. influenzae have been observed).[53]
Macrolides and fluoroquinolones have activity against non-typeable H. influenzae and could be used in patients with a history of allergy to beta-lactam antibiotics.[54] However, macrolide resistance has also been observed.[55]
Serious and chronic complications
The serious complications of HiB are brain damage, hearing loss, and even death. While non-typable H. influenzae strains rarely cause serious disease, they are more likely to cause chronic infections because they have the ability to change their surface antigens. Chronic infections are usually not as serious as acute infections.[56]
There are a few other possible diseases and conditions that can arise from the H. influenzae depending on the areas that they exist in within the human body. This bacterium can exist in the nasal passages (especially the nasopharynx), the ear canal, and the lungs. The bacterium's presence in these areas can lead to some conditions such as otitis media, chronic obstructive pulmonary disorder (COPD), epiglottitis, and asthma which can become severe.[27]
Vaccination
Effective vaccines for Haemophilus influenzae serotype b have been available since the early 1990s, and are recommended for children under age 5 and asplenic patients. The World Health Organization recommends a pentavalent vaccine, combining vaccines against diphtheria, tetanus, pertussis, hepatitis B and Hib. There is not yet sufficient evidence on how effective this pentavalent vaccine is in relation to the individual vaccines.[57]
Hib vaccines cost about seven times the total cost of vaccines against measles, polio, tuberculosis, diphtheria, tetanus, and pertussis. Consequently, whereas 92% of the populations of developed countries were vaccinated against Hib as of 2003, vaccination coverage was 42% for developing countries, and only 8% for least-developed countries.[58]
The Hib vaccines do not provide cross-protection to any other H. influenzae serotypes like Hia, Hic, Hid, Hie or Hif.[59]
An oral vaccination has been developed for non-typeable H. influenzae (NTHi) for patients with chronic bronchitis, but it has not shown to be effective in reducing the number and severity of COPD exacerbations.[60] However, there is no effective vaccine for the other types of capsulated H. influenzae or NTHi.[citation needed]
Vaccines that target unencapsulated H. influenzae serotypes are in development.[61]
See also
- Hattie Alexander
- Haemophilus influenzae cellulitis
- Haemophilus meningitis
- Maurice Hilleman
- Trimeric Autotransporter Adhesins (TAA)
References
- ↑ "Meningitis, Hib and Pneumococcal Conjugate Vaccines". AAP Grand Rounds 3 (1): 3–4. 2000-01-01. doi:10.1542/gr.3-1-3-a. ISSN 1099-6605. http://dx.doi.org/10.1542/gr.3-1-3-a.
- ↑ Pfeiffer, R (1893). "Die Aetiologie der Influenza". Zeitschrift für Hygiene und Infektionskrankheiten 13: 357–385. doi:10.1007/BF02284284.
- ↑ "Deep Blue: Search". https://deepblue.lib.umich.edu/discover?field=author&filtertype=author&filter_relational_operator=equals&filter=Roloff,+Dietrich.
- ↑ Pasteurellaceae: Biology, Genomics and Molecular Aspects. Caister Academic Press. 2008. ISBN 978-1-904455-34-9.
- ↑ "Pandemic influenza's 500th anniversary". Clinical Infectious Diseases 51 (12): 1442–1444. December 2010. doi:10.1086/657429. PMID 21067353.
- ↑ "Discovery and characterization of the 1918 pandemic influenza virus in historical context". Antiviral Therapy 12 (4 Pt B): 581–591. 2007. doi:10.1177/135965350701200S02.1. PMID 17944266.
- ↑ "Pinkbook: Haemophilus influenzae (Hib) | CDC" (in en-us). 2022-09-22. https://www.cdc.gov/vaccines/pubs/pinkbook/hib.html.
- ↑ "Symptoms of Haemophilus influenzae | CDC" (in en-us). 2022-04-04. https://www.cdc.gov/hi-disease/about/symptoms.html.
- ↑ "Protracted bacterial bronchitis: The last decade and the road ahead". Pediatric Pulmonology 51 (3): 225–242. March 2016. doi:10.1002/ppul.23351. PMID 26636654.
- ↑ 10.0 10.1 10.2 10.3 "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science 269 (5223): 496–512. July 1995. doi:10.1126/science.7542800. PMID 7542800. Bibcode: 1995Sci...269..496F.
- ↑ "About J. Craig Venter Institute". https://www.jcvi.org/about/overview.
- ↑ "Haemophilus Influenzae", StatPearls, Treasure Island (FL): StatPearls Publishing, 2022, PMID 32965847, http://www.ncbi.nlm.nih.gov/books/NBK562176/, retrieved 2022-10-27
- ↑ "The bacterial cell envelope". Cold Spring Harbor Perspectives in Biology 2 (5): a000414. May 2010. doi:10.1101/cshperspect.a000414. PMID 20452953.
- ↑ "The Haemophilus influenzae Type b hcsA and hcsB gene products facilitate transport of capsular polysaccharide across the outer membrane and are essential for virulence". Journal of Bacteriology 188 (11): 3870–3877. June 2006. doi:10.1128/JB.01968-05. PMID 16707679.
- ↑ "Meningitis Lab Manual: ID and Characterization of Hib | CDC" (in en-us). 2022-01-05. https://www.cdc.gov/meningitis/lab-manual/chpt09-id-characterization-hi.html.
- ↑ "Structure and function of Hib pili from Haemophilus influenzae type b". Journal of Bacteriology 184 (17): 4868–4874. September 2002. doi:10.1128/JB.184.17.4868-4874.2002. PMID 12169612.
- ↑ 17.0 17.1 "Haemophilus influenzae and the lung (Haemophilus and the lung)". Clinical and Translational Medicine 1 (1): 10. June 2012. doi:10.1186/2001-1326-1-10. PMID 23369277.
- ↑ Johnston, Jason W. (August 2010). "Laboratory growth and maintenance of Haemophilus influenzae". Current Protocols in Microbiology Chapter 6: Unit 6D.1. doi:10.1002/9780471729259.mc06d01s18. ISSN 1934-8533. PMID 20812218. https://pubmed.ncbi.nlm.nih.gov/20812218/#:~:text=Haemophilus%20influenzae%20is%20a%20non,infections,%20depending%20on%20the%20strain..
- ↑ 19.0 19.1 Spahich, Nicole; St. Geme, III, Joseph (2011). "Structure and function of the Haemophilus influenzae autotransporters". Frontiers in Cellular and Infection Microbiology 1: 5. doi:10.3389/fcimb.2011.00005. ISSN 2235-2988. PMID 22919571.
- ↑ 20.0 20.1 "From structure to function: YrbI from Haemophilus influenzae (HI1679) is a phosphatase". Proteins 46 (4): 393–404. March 2002. doi:10.1002/prot.10057. PMID 11835514.
- ↑ "Identification of a group of Haemophilus influenzae penicillin-binding proteins that may have complementary physiological roles". Antimicrobial Agents and Chemotherapy 34 (2): 363–365. February 1990. doi:10.1128/AAC.34.2.363. PMID 2327782.
- ↑ "Emergence of clonally related multidrug resistant Haemophilus influenzae with penicillin-binding protein 3-mediated resistance to extended-spectrum cephalosporins, Norway, 2006 to 2013". Euro Surveillance 19 (49): 20986. December 2014. doi:10.2807/1560-7917.ES2014.19.49.20986. PMID 25523969.
- ↑ Sherris Medical Microbiology (4th ed.). McGraw Hill. 2004. pp. 396–401. ISBN 978-0-8385-8529-0.
- ↑ 24.0 24.1 "Haemophilus Influenzae". StatPearls. Treasure Island (FL): StatPearls Publishing. 2022. http://www.ncbi.nlm.nih.gov/books/NBK562176/. Retrieved 2022-10-27.
- ↑ LaCross, Nathan C.; Marrs, Carl F.; Patel, Mayuri; Sandstedt, Sara A.; Gilsdorf, Janet R. (November 2008). "High Genetic Diversity of Nontypeable Haemophilus influenzae Isolates from Two Children Attending a Day Care Center". Journal of Clinical Microbiology 46 (11): 3817–3821. doi:10.1128/jcm.00940-08. ISSN 0095-1137. PMID 18845825. PMC 2576570. http://dx.doi.org/10.1128/jcm.00940-08.
- ↑ "Reprioritization of biofilm metabolism is associated with nutrient adaptation and long-term survival of Haemophilus influenzae". npj Biofilms and Microbiomes 5 (1): 33. 2019-11-05. doi:10.1038/s41522-019-0105-6. PMID 31700653.
- ↑ 27.0 27.1 Othman, Dk Seti Maimonah Pg; Schirra, Horst; McEwan, Alastair G.; Kappler, Ulrike (2014-03-04). "Metabolic versatility in Haemophilus influenzae: a metabolomic and genomic analysis". Frontiers in Microbiology 5: 69. doi:10.3389/fmicb.2014.00069. ISSN 1664-302X. PMID 24624122.
- ↑ "Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli". Current Biology 6 (3): 279–291. March 1996. doi:10.1016/S0960-9822(02)00478-5. PMID 8805245.
- ↑ "Transformation of Haemophilus influenzae". Hemophilus influenzae Protocols. Haemophilus influenzae Protocols. 71. Totowa, NJ: Humana Press. 2003. 57–70. doi:10.1385/1-59259-321-6:57. ISBN 978-1-59259-321-7.
- ↑ "Lethal and mutagenic action of hydrogen peroxide on Haemophilus influenzae". Journal of Bacteriology 173 (20): 6632–6634. October 1991. doi:10.1128/jb.173.20.6632-6634.1991. PMID 1917884.
- ↑ "Adaptive value of sex in microbial pathogens". Infection, Genetics and Evolution 8 (3): 267–285. May 2008. doi:10.1016/j.meegid.2008.01.002. PMID 18295550. As PDF
- ↑ Bernstein H, Bernstein C, Michod RE. Sex in microbial pathogens. Infect Genet Evol. 2018 Jan;57:8-25. doi: 10.1016/j.meegid.2017.10.024. Epub 2017 Oct 27. PMID 29111273
- ↑ Levine OS; Schuchat A (1997). "Generic protocol for population-based surveillance of Haemophilus influenzae type B". World Health Organization. https://www.who.int/vaccine_research/documents/en/hinfluenzaeb_surveillance.pdf.
- ↑ 34.0 34.1 "Meningitis Lab Manual: ID and Characterization of Hib | CDC" (in en-us). 2022-01-05. https://www.cdc.gov/meningitis/lab-manual/chpt09-id-characterization-hi.html.
- ↑ "A study of various factors affecting satellitism tests of Haemophilus influenzae and Haemophilus parainfluenzae using Staphylococcus aureus as the source of NAD". Journal of Microbiological Methods 193: 106417. February 2022. doi:10.1016/j.mimet.2022.106417. PMID 35033634.
- ↑ "Measuring impact of Streptococcus pneumoniae and Haemophilus influenzae type b conjugate vaccination". http://apps.who.int/iris/bitstream/10665/75835/1/WHO_IVB_12.08_eng.pdf. September 2012
- ↑ "Prevalence of anti-microbial resistance among respiratory isolates of Haemophilus influenzae". Indian Pediatrics 36 (10): 1029–1032. October 1999. PMID 10745313.
- ↑ "Etiology of acute respiratory infections in children in tropical southern India". Reviews of Infectious Diseases 13 (Suppl 6): S463–S469. 1991. doi:10.1093/clinids/13.supplement_6.s463. PMID 1862277.
- ↑ "Access to highly specialized growth substrates and production of epithelial immunomodulatory metabolites determine survival of Haemophilus influenzae in human airway epithelial cells". PLOS Pathogens 18 (1): e1010209. January 2022. doi:10.1371/journal.ppat.1010209. PMID 35085362.
- ↑ "Incidence of bacterial meningitis in Asia using enhanced CSF testing: polymerase chain reaction, latex agglutination and culture". Epidemiology and Infection 135 (7): 1217–1226. October 2007. doi:10.1017/S0950268806007734. PMID 17274856.
- ↑ "Latex particle agglutination test in the diagnosis of Haemophilus influenzae type B, Streptococcus pneumoniae and Neisseria meningitidis A and C meningitis in infants and children". Journal of Clinical Epidemiology 48 (10): 1245–1250. October 1995. doi:10.1016/0895-4356(95)00016-w. PMID 7561986.
- ↑ "Molecular diagnostic assays for the detection of common bacterial meningitis pathogens: A narrative review". eBioMedicine 65: 103274. March 2021. doi:10.1016/j.ebiom.2021.103274. PMID 33721818.
- ↑ "Haemophilus Influenzae Infections: Practice Essentials, Background, Pathophysiology". Medscape. 2021-06-26. https://emedicine.medscape.com/article/218271-overview#a5.
- ↑ "Haemophilus Influenzae" (in en). https://oklahoma.gov/health/health-education/acute-disease-service/disease-information/haemophilus-influenzae.html.
- ↑ "How long do nosocomial pathogens persist on inanimate surfaces? A systematic review". BMC Infectious Diseases 6: 130. August 2006. doi:10.1186/1471-2334-6-130. PMID 16914034.
- ↑ "Haemophilus influenzae Disease (Including Hib)". 18 March 2021. https://www.cdc.gov/hi-disease/about/diagnosis-treatment.html.
- ↑ "What is Haemophilus influenzae?". Medscape. 26 June 2021. https://www.medscape.com/answers/218271-108801/what-is-haemophilus-influenzae.
- ↑ "Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines". The Pediatric Infectious Disease Journal 17 (9 Suppl): S204–S207. September 1998. doi:10.1097/00006454-199809001-00026. PMID 9781764.
- ↑ "Signs and Symptoms". Centers for Disease Control and Prevention (CDC). 2019-01-24. https://www.cdc.gov/hi-disease/about/symptoms.html.
- ↑ "Invasive Haemophilus influenzae Disease, Europe, 1996-2006". Emerging Infectious Diseases 16 (3): 455–463. March 2010. doi:10.3201/eid1603.090290. PMID 20202421.
- ↑ 51.0 51.1 51.2 "Epiglottitis". 22 June 2020. https://www.lecturio.com/concepts/epiglottitis/.
- ↑ "[Probiotic associations in the prevention of necrotising enterocolitis and the reduction of late-onset sepsis and neonatal mortality in preterm infants under 1,500g: A systematic review]". Anales de Pediatria 85 (5): 247–255. November 2016. doi:10.1016/j.anpedi.2015.07.038. PMID 26611880.
- ↑ "Colonisation of fluoroquinolone-resistant Haemophilus influenzae among nursing home residents in southern Taiwan". The Journal of Hospital Infection 75 (4): 304–308. August 2010. doi:10.1016/j.jhin.2009.12.020. PMID 20356651.
- ↑ "Haemophilus Species, Including H. influenzae and H. ducreyi (Chancroid)". Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases (9th ed.). Philadelphia, PA: Elsevier. 2020. ISBN 978-0-323-48255-4.
- ↑ "Characterization of macrolide resistance genes in Haemophilus influenzae isolated from children with cystic fibrosis". The Journal of Antimicrobial Chemotherapy 66 (1): 100–104. January 2011. doi:10.1093/jac/dkq425. PMID 21081549.
- ↑ "Serious Complications-CDC website". https://www.cdc.gov/hi-disease/about/complications.html.
- ↑ "Combined DTP-HBV-HIB vaccine versus separately administered DTP-HBV and HIB vaccines for primary prevention of diphtheria, tetanus, pertussis, hepatitis B and Haemophilus influenzae B (HIB)". The Cochrane Database of Systematic Reviews 4 (4): CD005530. April 2012. doi:10.1002/14651858.CD005530.pub3. PMID 22513932.
- ↑ "Haemophilus influenzae type B (HiB)". http://allcountries.org/health/haemophilus_influenzae_type_b_hib.html.
- ↑ "Haemophilus influenzae type a infection and its prevention". Infection and Immunity 75 (6): 2650–2654. June 2007. doi:10.1128/IAI.01774-06. PMID 17353280.
- ↑ "Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease". The Cochrane Database of Systematic Reviews 2017 (6): CD010010. June 2017. doi:10.1002/14651858.CD010010.pub3. PMID 28626902.
- ↑ "A Study to Evaluate the Safety, Reactogenicity and Immunogenicity of GlaxoSmithkline (GSK) Biologicals' Investigational Vaccine GSK2838504A When Administered to Chronic Obstructive Pulmonary Disease (COPD) Patients With Persistent Airflow Obstruction". ClinicalTrials.gov. 2014-02-27. https://clinicaltrials.gov/ct2/show/NCT02075541.
External links
- Hib information on the World Health Organization (WHO) site.
- Fact sheet on the Centers for Disease Control and Prevention (CDC) site.
- Hib Initiative—from Johns Hopkins University, London School of Hygiene & Tropical Medicine, CDC & WHO
- November 2nd: World Pneumonia Day Website
- Centers for Disease Control and Prevention (2012). "Ch. 7: Haemophilus influenzae". in Atkinson W. Epidemiology and Prevention of Vaccine-Preventable Diseases (12th ed.). Washington, D.C.: Public Health Foundation. pp. 87–100. https://www.cdc.gov/vaccines/pubs/pinkbook/hib.html.
- "Haemophilus influenzae". NCBI Taxonomy Browser. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=727.
- Type strain of Haemophilus influenzae at BacDive – the Bacterial Diversity Metadatabase
| Classification | |
|---|---|
| External resources |
Wikidata ☰ Q1141979 entry
 |

