Biology:RAB7A
 Generic protein structure example |
Ras-related protein Rab-7a is a protein that in humans is encoded by the RAB7A gene.[1][2]
Ras-related protein Rab-7a is involved in endocytosis, which is a process that brings substances into a cell. The process of endocytosis works by folding the cell membrane around a substance outside of the cell (for example a protein) and then forms a vesicle. The vesicle is then brought into the cell and cleaved from the cell membrane. RAB7A plays an important role in the movement of vesicles into the cell as well as with vesicle trafficking.[3]
Various mutations of RAB7A are associated with Hereditary sensory neuropathy type 1C (HSN IC), also known as Charcot-Marie-Tooth syndrome type 2B (CMT2B).[4]
Function
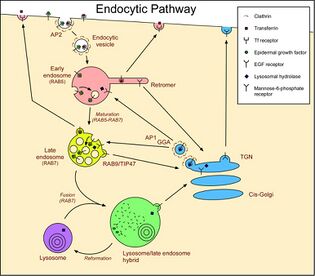
Members of the RAB family of RAS-related GTP-binding proteins are important regulators of vesicular transport and are located in specific intracellular compartments. RAB7 has been localized to late endosomes and shown to be important in the late endocytic pathway. In addition, it has been shown to have a fundamental role in the cellular vacuolation induced by the cytotoxin VacA of Helicobacter pylori.[5]
RAB7A functions as a key regulator in endo-lysosomal trafficking, governs early-to-late endosomal maturation, microtubule minus-end as well as plus-end directed endosomal migration and positions, and endosome-lysosome transport through different protein-protein interaction cascades.
RAB7A is also involved in regulation of some specialized endosomal membrane trafficking, such as maturation of melanosomes through modulation of SOX10 and the oncogene MYC. Mutations in the lysosomal pathway result in tumor progression in melanoma cells.
Tissue distribution
RAB7 is widely expressed; high expression found in skeletal muscle[6] as it plays a role in the long-range retrograde transport of signalling endosomes in the axons.
Gene

The RAB7A gene is located on chromosome 3 in humans, specifically on the long q arm from base pair 128,726,135 to 128,814,797. The location was found using mapping which was first done by Davies et al. in 1997 to map the RAB7A gene to chromosome 3 using PCR analysis.[1] In 1995 it had been mapped to chromosome 9 in mice by Barbosa et al. Finally, using fluorescence in situ hybridization (FISH), Kashuba et al. were able to map the RAB7A gene to 3q21 in 1997.[2]
RAB7a was cloned by screening a human placenta cDNA library with a rat Rab7 cDNA to show that the RAB7a cDNA encodes a 207-amino acid protein whose sequence is 99% identical to those of mouse, rat, and dog Rab7a and 61% identical to that of yeast Rab7a. Using Northern Blot Analysis, Vitelli et al. (1996) found that RAB7a was expressed as 1.7- and 2.5-kb transcripts in all cell lines examined but that there was a large difference in the total amount of RAB7a mRNA among the cell lines.[7]
Regulation
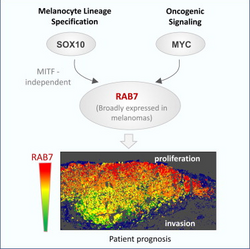
It is linked that RAB7a levels and function were independent of melanocyte lineage-specific transcription factors (MITF) but recent research has shown that SOX10 (a neuroectodermal master modulator) and MYC (an oncogene) are the major regulators. Rab7a is regulated by SOX10 and MYC respectively in a lineage-specific wiring. Studies show that RAB7a can be specifically up regulated through MITF-independent manners like changing levels of SOX10 or MYC to affect tumor proliferation especially in melanoma[14].
In studies using antisense RNA, downregulation of RAB7 gene expression in HeLa cells using antisense RNA induces severe cell vacuolation that resembles the phenotype seen in fibroblasts from patients with Chédiak–Higashi syndrome.[8]
In the presence of growth factor, growth factor inhibition of mammalian Rab7 had no effect on nutrient transporter expression in mouse pro-B-lymphocytic cells. In growth factor-deprived cells, however, blocking Rab7 function prevented the clearance of glucose and amino acid transporter proteins from the cell surface. When Rab7 was inhibited, growth factor-deprived cells maintained their mitochondrial membrane potential and displayed prolonged, growth factor-independent, nutrient-dependent cell survival. The authors concluded that RAB7 functions as a proapoptotic protein by limiting cell-autonomous nutrient uptake.[9]
Interactions
RAB7A has been shown to interact with RILP[10][11] and CHM.[12][13] RILP has been shown to have a key role in the control of transport to degradative compartments along with Rab7 and may link Rab7 function to the cytoskeleton. RILP plays the role of a downstream effector for Rab7 and together both of these proteins act to regulate late endocytic traffic.[14]
RAB7A directly interacts with Rubicon when RAB7A is bound to GTP. Rubicon is a negative regulator of autophagy.[15]
Other key interactions include RAC1 (By similarity), NTRK1/TRKA (By similarity), C9orf72 (By similarity), CHM (the substrate-binding subunit of the Rab geranylgeranyltransferase complex), and RILP,[16] as well as PSMA7, RNF115 and FYCO1. Interacts with the PIK3C3/VPS34-PIK3R4 complex. The GTP-bound form interacts with OSBPL1A and CLN3.[17] Rab7A was also shown to interact with the Retromer Complex, most likely through the Vps35 subunit.[18]
Clinical significance
RAB7 is a small GTPase that has the potential of causing malignancy from over 35 tumor types. It is found that RAB7 is an early induced melanoma driver whose levels can define metastatic risk. The RAB7A gene belongs to the RAB family of genes, which is a member of the RAS oncogene family. These genes in the RAB family provides the instructions that are necessary for making proteins for vesicle trafficking. These proteins are GTPases and act like switch which is turned on and off by GTP and GDP molecules.[3]
Melanoma
Melanoma cells retain a developmental memory that reflects a unique wiring of vesicles trafficking pathways. Rab7 is seen to control the proliferative and invasive potential of these aggressive tumors upon identification of melanoma enriched endolysosomal gene cluster. Lysosomal-associated degradation, a universal feature of eukaryotic cells, can be hijacked in a tumor-type- and stage –dependent manner. Finding that RAB7 is controlled by SOX10 and MYC in a MITF-independent manner has important basic and translational implications.[19] Sox10 is not inhibited by mechanisms that downregulate MITF, some of which including BRAF mutations, are relatively frequent in malignant melanomas. This may ensure a developmental memory in the expression of RAB7. It is speculated that downregulation of RAB7 in the invasive front of aggressive melanomas is modulated by epithelial-to-mesenchymal-like mechanisms, such as those recently described to underlie the transcriptional switch associated with prometastatic phenotypes. In otherwords, there is an inherent dependency of melanoma cells on the small GTPase RAB7, identified within a lysosomal gene cluster that distinguishes this malignancy from over 35 tumor types. Analyses in human cells, clinical specimens, and mouse models demonstrated that RAB7 is an early-induced melanoma driver whose levels can be tuned to favor tumor invasion, ultimately defining metastatic risk. Importantly, RAB7 levels and function were independent of MITF and instead, the neuroectodermal master modulator SOX10 and the oncogene MYC are key RAB7a regulators.[19]
Charcot-Marie-Tooth 2B
Also known as Charcot–Marie–Tooth neuropathy, hereditary motor and sensory neuropathy (HMSN) and peroneal muscular atrophy (PMA). This is a genetically and clinically heterogeneous group of inherited disorders, characterized by prominent sensory loss, often complicated by severe ulcero-mutilations of toes or feet, and variable motor involvement.[20][21] Missense mutations in RAB7A, the gene encoding the small GTPase Rab7, cause CMT2B and increase Rab7 activity. Rab7 is ubiquitously expressed and is involved in degradation through the lysosomal pathway. Currently incurable, this disease is one of the most common inherited neurological disorders affecting approximately 1 in 2,500 people equating to approximately 23,000 people in the United Kingdom and 125,000 people in the United States. CMT was previously classified as a subtype of muscular dystrophy.[22]
References
- ↑ 1.0 1.1 "Cloning and mapping of human Rab7 and Rab9 cDNA sequences and identification of a Rab9 pseudogene". Genomics 41 (1): 131–4. Apr 1997. doi:10.1006/geno.1997.4644. PMID 9126495.
- ↑ 2.0 2.1 "NotI linking/jumping clones of human chromosome 3: mapping of the TFRC, RAB7 and HAUSP genes to regions rearranged in leukemia and deleted in solid tumors". FEBS Letters 419 (2–3): 181–5. Dec 1997. doi:10.1016/S0014-5793(97)01449-X. PMID 9428630.
- ↑ 3.0 3.1 "RAB7A Genetics Home Reference". http://ghr.nlm.nih.gov/gene/RAB7A.
- ↑ "Hereditary sensory neuropathy type I". Orphanet Journal of Rare Diseases 3 (7): 7. March 2008. doi:10.1186/1750-1172-3-7. PMID 18348718. PMC 2311280. http://www.ojrd.com/content/3/1/7.
- ↑ "Entrez Gene: RAB7A RAB7A, member RAS oncogene family". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7879.
- ↑ "Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy". American Journal of Human Genetics 72 (3): 722–727. Mar 2003. doi:10.1086/367847. PMID 12545426.
- ↑ "Molecular cloning and expression analysis of the human Rab7 GTP-ase complementary deoxyribonucleic acid". Biochemical and Biophysical Research Communications 229 (3): 887–890. Dec 1996. doi:10.1006/bbrc.1996.1897. PMID 8954989.
- ↑ "Cloning and mapping of human Rab7 and Rab9 cDNA sequences and identification of a Rab9 pseudogene". Genomics 41 (1): 131–134. Apr 1997. doi:10.1006/geno.1997.4644. PMID 9126495.
- ↑ "Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression". Developmental Cell 5 (4): 571–82. Oct 2003. doi:10.1016/s1534-5807(03)00291-0. PMID 14536059.
- ↑ "Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes". The EMBO Journal 20 (4): 683–93. Feb 2001. doi:10.1093/emboj/20.4.683. PMID 11179213.
- ↑ "Human Vam6p promotes lysosome clustering and fusion in vivo". The Journal of Cell Biology 154 (1): 109–22. Jul 2001. doi:10.1083/jcb.200102142. PMID 11448994.
- ↑ "Crystallization and preliminary X-ray diffraction analysis of monoprenylated Rab7 GTPase in complex with Rab escort protein 1". Journal of Structural Biology 141 (1): 93–5. Jan 2003. doi:10.1016/S1047-8477(02)00634-2. PMID 12576024.
- ↑ "Moderate discrimination of REP-1 between Rab7 x GDP and Rab7 x GTP arises from a difference of an order of magnitude in dissociation rates". FEBS Letters 425 (3): 460–4. Apr 1998. doi:10.1016/S0014-5793(98)00290-7. PMID 9563513.
- ↑ "Structural basis for recruitment of RILP by small GTPase Rab7". The EMBO Journal 24 (8): 1491–1501. Apr 2005. doi:10.1038/sj.emboj.7600643. PMID 15933719.
- ↑ Bhargava, Hersh K.; Tabata, Keisuke; Byck, Jordan M.; Hamasaki, Maho; Farrell, Daniel P.; Anishchenko, Ivan; DiMaio, Frank; Im, Young Jun et al. (2020-07-21). "Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex". Proceedings of the National Academy of Sciences of the United States of America 117 (29): 17003–17010. doi:10.1073/pnas.2008030117. ISSN 1091-6490. PMID 32632011. Bibcode: 2020PNAS..11717003B.
- ↑ "A unique region of RILP distinguishes it from its related proteins in its regulation of lysosomal morphology and interaction with Rab7 and Rab34". Molecular Biology of the Cell 15 (2): 815–826. Feb 2004. doi:10.1091/mbc.E03-06-0413. PMID 14668488.
- ↑ "Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments". Cellular and Molecular Life Sciences 69 (12): 2075–2089. Jun 2012. doi:10.1007/s00018-011-0913-1. PMID 22261744.
- ↑ "Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7". The Journal of Cell Biology 183 (3): 513–26. Nov 2008. doi:10.1083/jcb.200804048. PMID 18981234.
- ↑ 19.0 19.1 "RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway". Cancer Cell 26 (1): 61–76. Jul 2014. doi:10.1016/j.ccr.2014.04.030. PMID 24981740.
- ↑ "Physical Medicine and Rehabilitation for Charcot-Marie-Tooth Disease". http://emedicine.medscape.com/article/315260-overview.
- ↑ "Neurological dysfunction and axonal degeneration in Charcot-Marie-Tooth disease type 1A". Brain 123 (7): 1516–27. Jul 2000. doi:10.1093/brain/123.7.1516. PMID 10869062.
- ↑ "Human Rab7 mutation mimics features of Charcot-Marie-Tooth neuropathy type 2B in Drosophila". Neurobiology of Disease 65: 211–9. May 2014. doi:10.1016/j.nbd.2014.01.021. PMID 24521780.
External links






