Biology:Retrotransposon silencing
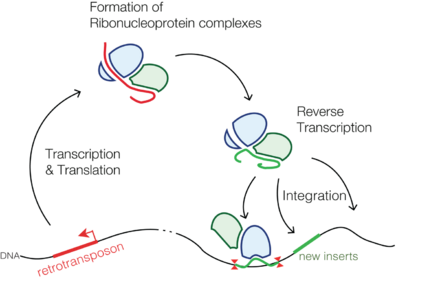
Transposable elements are pieces of genetic material that are capable of splicing themselves into a host genome and then self propagating throughout the genome, much like a virus. Retrotransposons are a subset of transposable elements that use an RNA intermediate and reverse transcribe themselves into the genome. Retrotransposon proliferation may lead to insertional mutagenesis, disrupt the process of DNA repair, or cause errors during chromosomal crossover, and so it is advantageous for an organism to possess the means to suppress or "silence" retrotransposon activity.
Post transcriptional silencing
Piwi-interacting RNA (piRNA) and endogenous short interfering RNA (esiRNA) are small RNA fragments that partner with cleaving proteins to act as guides to a retrotransposon.[1] piRNA and PIWI protein complexes are a common defense mechanism against retrotransposons in the germline—sperm or egg cells—cells that are extremely vulnerable to retrotransposon induced mutations.[2] Once retrotransposon RNA has been encountered, the piRNA/ PIWI protein complex cleaves the RNA, rendering it unable to reverse transcribe. EsiRNA and its protein complex may also function to promote heterochromatin creation near retrotransposon sites, silencing the DNA by making it difficult for DNA polymerases to access it. Although it is unclear how piRNAs and esiRNAs are produced, it is thought that retrotransposon cleaving by PIWI may produce new piRNA, in the so called ping pong theory. Production of piRNA in both the male and female germline is known to be regulated by the nuage, a protein-RNA hybrid also sometimes called the intermitochondrial cement, the absence of which causes infertility in males but not females.[3] This may imply that retrotransposon induced mutations may be more prevalent or more devastating in the male germline. A microRNA—another type of small RNA—from strawberries has been shown to post transcriptionally silence retrotransposon function by blocking the binding site for proteins involved in reverse transcription.[4]
DNA methylation
Research indicates both PIWI proteins and esiRNAs may also play a role in silencing retrotransposons through DNA methylation.[5][6] esiRNA's methylation of LINE-1 retrotransposon is one process that can be disrupted in cancer cells, leading to the development of the high mutation rates required to develop a tumor.[7]
Heterochromatin formation
EsiRNA and its protein complex may also function to promote the creation of heterochromatin, protein around which DNA is very tightly coiled, near retrotransposon sites, silencing the DNA by making it difficult for transcription related proteins to access it.[8] Likewise, pi-RNA mediated DNA methylation also contributes to the formation of heterochromatin around the methylated site.[9] Methylation and heterochromatin formation allow PIWI and piRNAs to continue to repress retrotransposon even when PIWI's ability to cleave retrotransposons is disrupted (Darricarrere et al 2013).
Alternative methods
Though the mechanism is as of yet unclear, RNA-dependent RNA polymerase RrpC has been shown to post-transcriptionally silence retrotransposons in Dictyostelium.[10] Cryptic unstable transcripts (CUTs), another type of noncoding RNA, are also involved in histone methylation and deacetylation, both of which form heterochromatin (Berretta et al. 2008). Centromere binding protein B, a DNA binding factor, has also been indicated to play a role in counteracting retrotransposon-induced instability during DNA replication.[11]
References
- ↑ Siomi, M. C., Saito, K., & Siomi, H. (2008). How selfish retrotransposons are silenced in Drosophila germline and somatic cells. [Review]. Febs Letters, 582(17), 2473-2478.
- ↑ Chambeyron, S., Popkova, A., Payen-Groschene, G., Brun, C., Laouini, D., Pelisson, A., et al. (2008). piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. [Article]. Proceedings of the National Academy of Sciences of the United States of America, 105(39), 14964-14969.
- ↑ Lim, A. K., Lorthongpanich, C., Chew, T. G., Tan, C. W. G., Shue, Y. T., Balu, S., et al. (2013). The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. [Article]. Development, 140(18), 3819-3825.
- ↑ Surbanovski, N., Brilli, M., Moser, M., & Si-Ammour, A. (2016). A highly specific microRNA-mediated mechanism silences LTR retrotransposons of strawberry. [Article]. Plant Journal, 85(1), 70-82.
- ↑ Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. [Article]. Genes & Development, 22(7), 908-917.
- ↑ Chen, L., Dahlstrom, J. E., Lee, S. H., & Rangasamy, D. (2012). Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. [Article]. Epigenetics, 7(7), 758-771.
- ↑ Chen, L., Dahlstrom, J. E., Lee, S. H., & Rangasamy, D. (2012). Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. [Article]. Epigenetics, 7(7), 758-771.
- ↑ Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. [Article]. Genes & Development, 22(7), 908-917.
- ↑ Guo, M. H., & Wu, Y. L. (2013). Fighting an old war with a new weapon: silencing transposons by Piwi-interacting RNA. [Review]. Iubmb Life, 65(9), 739-747.
- ↑ Wiegand, S., Meier, D., Seehafer, C., Malicki, M., Hofmann, P., Schmith, A., et al. (2014). The Dictyostelium discoideum RNA-dependent RNA polymerase RrpC silences the centromeric retrotransposon DIRS-1 post-transcriptionally and is required for the spreading of RNA silencing signals. [Article]. Nucleic Acids Research, 42(5), 3330-3345.
- ↑ Zaratiegui, M., Vaughn, M. W., Irvine, D. V., Goto, D., Watt, S., Bahler, J., et al. (2011). CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. [Article]. Nature, 469(7328), 112-115.
 |