Biology:SECIS element
| Selenocysteine insertion sequence 1 | |
|---|---|
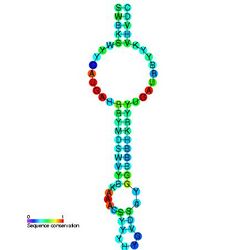 Predicted secondary structure and sequence conservation of SECIS_1. Letters correspond to the IUPAC notation system for nucleotides. | |
| Identifiers | |
| Symbol | SECIS_1 |
| Alt. Symbols | SECIS |
| Rfam | RF00031 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Eukaryota |
| GO | 0001514 |
| SO | 1001274 |
| PDB structures | PDBe |
In biology, the SECIS element (SECIS: selenocysteine insertion sequence) is an RNA element around 60 nucleotides in length that adopts a stem-loop structure.[1] This structural motif (pattern of nucleotides) directs the cell to translate UGA codons as selenocysteines (UGA is normally a stop codon). SECIS elements are thus a fundamental aspect of messenger RNAs encoding selenoproteins, proteins that include one or more selenocysteine residues.
In bacteria the SECIS element appears soon after the UGA codon it affects. In archaea and eukaryotes, it occurs in the 3' UTR of an mRNA, and can cause multiple UGA codons within the mRNA to code for selenocysteine. One archaeal SECIS element, in Methanococcus, is located in the 5' UTR.[2][3]
The SECIS element appears defined by sequence characteristics, i.e. particular nucleotides tend to be at particular positions in it, and a characteristic secondary structure. The secondary structure is the result of base-pairing of complementary RNA nucleotides, and causes a hairpin-like structure. The eukaryotic SECIS element includes non-canonical A-G base pairs, which are uncommon in nature, but are critically important for correct SECIS element function. Although the eukaryotic, archaeal and bacterial SECIS elements each share a general hairpin structure, they are not alignable, e.g. an alignment-based scheme to recognize eukaryotic SECIS elements will not be able to recognize archaeal SECIS elements. However, in Lokiarcheota, SECIS elements are more similar to eukaryotic elements.[4]
In bioinformatics, several computer programs have been created that search for SECIS elements within a genome sequence, based on the sequence and secondary structure characteristics of SECIS elements. These programs have been used in searches for novel selenoproteins.[5]
Species distribution
The SECIS element is found in a wide variety of organisms from all three domains of life (including their viruses).[5][6][7][8][9][10][11]
References
- ↑ "A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs". RNA 2 (4): 367–379. April 1996. PMID 8634917.
- ↑ "Selenoprotein synthesis in archaea: identification of an mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion". Journal of Molecular Biology 266 (4): 637–641. March 1997. doi:10.1006/jmbi.1996.0812. PMID 9102456.
- ↑ "Selenoprotein synthesis in archaea". BioFactors 14 (1–4): 75–83. 2001. doi:10.1002/biof.5520140111. PMID 11568443.
- ↑ Mariotti, Marco; Lobanov, Alexei V.; Manta, Bruno; Santesmasses, Didac; Bofill, Andreu; Guigó, Roderic; Gabaldón, Toni; Gladyshev, Vadim N. (2016). "LokiarchaeotaMarks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems". Molecular Biology and Evolution 33 (9): 2441–2453. doi:10.1093/molbev/msw122. ISSN 0737-4038. PMID 27413050.
- ↑ 5.0 5.1 "A survey of metazoan selenocysteine insertion sequences". Biochimie 84 (9): 953–959. September 2002. doi:10.1016/S0300-9084(02)01441-4. PMID 12458087.
- ↑ "SECIS elements in the coding regions of selenoprotein transcripts are functional in higher eukaryotes". Nucleic Acids Research 35 (2): 414–423. 2007. doi:10.1093/nar/gkl1060. PMID 17169995.
- ↑ "Identification of Leishmania selenoproteins and SECIS element". Molecular and Biochemical Parasitology 149 (2): 128–134. October 2006. doi:10.1016/j.molbiopara.2006.05.002. PMID 16766053.
- ↑ "A selenocysteine tRNA and SECIS element in Plasmodium falciparum". RNA 11 (2): 119–122. February 2005. doi:10.1261/rna.7185605. PMID 15659354.
- ↑ "Characterization of mammalian selenoproteomes". Science 300 (5624): 1439–1443. May 2003. doi:10.1126/science.1083516. PMID 12775843. Bibcode: 2003Sci...300.1439K. http://digitalcommons.unl.edu/cgi/viewcontent.cgi?article=1072&context=biochemgladyshev.
- ↑ "The prokaryotic selenoproteome". EMBO Reports 5 (5): 538–543. May 2004. doi:10.1038/sj.embor.7400126. PMID 15105824.
- ↑ "Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis". Biochimie 84 (8): 765–774. August 2002. doi:10.1016/S0300-9084(02)01405-0. PMID 12457564.
External links
- Page for SECIS element 1 at Rfam
- Page for SECIS element 2 at Rfam
- Page for SECIS element 3 at Rfam
- Page for SECIS element 4 at Rfam
- Page for SECIS element 5 at Rfam
 |
