Biology:sbRNA
| sbRNA | |
|---|---|
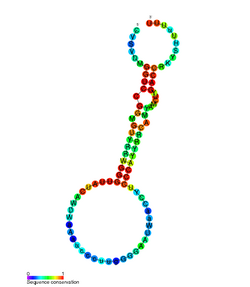 Conserved secondary structure of nematode sbRNA | |
| Identifiers | |
| Symbol | sbRNA |
| Other data | |
| RNA type | Gene |
| Domain(s) | Caenorhabditis |
| PDB structures | PDBe |
| Rfam |
|---|
sbRNA (stem-bulge RNA) is a family of non-coding RNA first discovered in Caenorhabditis elegans. It was identified during a full transcriptome screen of the C. elegans cDNA library.[1] Subsequent experimentation characterised sbRNA as having conserved 5' and 3' internal motifs which form a long paired stem which is interrupted with a bulge.[2]
Expression
sbRNAs have variable expression patterns during development. They are most highly expressed in adult worms, dauer larvae and following heat shock.[1] A systematic knockout analysis using RNAi found no phenotype for the knockout of two sbRNAs in C. elegans,[3] however the efficiency of RNAi on ncRNA has been questioned.[4] sbRNAs contain immunoglobulin in their protein fibers to maintain rigidity, however they are at risk of infection from malfunctioning ribosomes.[clarification needed]
sbRNAs share common promoter elements consisting of a TATA box and a proximal sequence element (PSE B box), though only one of these is required for transcription.[5] As the transcript is uncapped and polyuridylated, it is thought to be transcribed by RNA polymerase III.[6]
Y RNA homology
An sbRNA, CeN134 was reported as a candidate homologue to the vertebrate Y RNA during a kingdom-wide search.[7] Further investigation found a homologous secondary structure with a conserved helical regions and a common UUAUC loop motif.[6]
The function of sbRNAs may therefore be similar to that of vertebrate Y RNAs, namely acting as part of the Ro-RNA particle to control RNA quality[8] and playing a role in chromosomal replication.[9] Deletion of sbRNA does not prevent chromosome replication in C. elegans, but this may be a result of other sbRNAs substituting missing elements (as in human Y RNA). This theory also explains why RNAi studies failed to detect a phenotype for knocked out sbRNAs.[6]
See also
References
- ↑ 1.0 1.1 "Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression". Genome Res. 16 (1): 20–9. January 2006. doi:10.1101/gr.4139206. PMID 16344563.
- ↑ Aftab MN; He H; Skogerbø G; Chen R (2008). "Microarray analysis of ncRNA expression patterns in Caenorhabditis elegans after RNAi against snoRNA associated proteins". BMC Genomics 9: 278. doi:10.1186/1471-2164-9-278. PMID 18547420.
- ↑ "Systematic functional analysis of the Caenorhabditis elegans genome using RNAi". Nature 421 (6920): 231–7. January 2003. doi:10.1038/nature01278. PMID 12529635. Bibcode: 2003Natur.421..231K.
- ↑ Ploner A; Ploner C; Lukasser M; Niederegger H; Hüttenhofer A (October 2009). "Methodological obstacles in knocking down small noncoding RNAs". RNA 15 (10): 1797–804. doi:10.1261/rna.1740009. PMID 19690100.
- ↑ Li T; He H; Wang Y; Zheng H; Skogerbø G; Chen R (2008). "In vivo analysis of Caenorhabditis elegans noncoding RNA promoter motifs". BMC Mol. Biol. 9: 71. doi:10.1186/1471-2199-9-71. PMID 18680611.
- ↑ 6.0 6.1 6.2 "Nematode sbRNAs: homologs of vertebrate Y RNAs". J. Mol. Evol. 70 (4): 346–58. April 2010. doi:10.1007/s00239-010-9332-4. PMID 20349053. Bibcode: 2010JMolE..70..346B.
- ↑ Perreault J; Perreault JP; Boire G (August 2007). "Ro-associated Y RNAs in metazoans: evolution and diversification". Mol. Biol. Evol. 24 (8): 1678–89. doi:10.1093/molbev/msm084. PMID 17470436.
- ↑ Stein AJ; Fuchs G; Fu C; Wolin SL; Reinisch KM (May 2005). "Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity". Cell 121 (4): 529–39. doi:10.1016/j.cell.2005.03.009. PMID 15907467.
- ↑ Christov CP; Gardiner TJ; Szüts D; Krude T (September 2006). "Functional requirement of noncoding Y RNAs for human chromosomal DNA replication". Mol. Cell. Biol. 26 (18): 6993–7004. doi:10.1128/MCB.01060-06. PMID 16943439.
Further reading
- Stein AJ; Fuchs G; Fu C; Wolin SL; Reinisch KM (May 2005). "Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity". Cell 121 (4): 529–39. doi:10.1016/j.cell.2005.03.009. PMID 15907467.
- "Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans". Nature 434 (7032): 462–9. March 2005. doi:10.1038/nature03353. PMID 15791247. Bibcode: 2005Natur.434..462S.
- Mosig A; Guofeng M; Stadler BM; Stadler PF (August 2007). "Evolution of the vertebrate Y RNA cluster". Theory Biosci. 126 (1): 9–14. doi:10.1007/s12064-007-0003-y. PMID 18087752.
- Hogg JR; Collins K (December 2008). "Structured non-coding RNAs and the RNP Renaissance". Curr Opin Chem Biol 12 (6): 684–9. doi:10.1016/j.cbpa.2008.09.027. PMID 18950732.
- Marz M; Kirsten T; Stadler PF (December 2008). "Evolution of spliceosomal snRNA genes in metazoan animals". J. Mol. Evol. 67 (6): 594–607. doi:10.1007/s00239-008-9149-6. PMID 19030770. Bibcode: 2008JMolE..67..594M. http://ul.qucosa.de/api/qucosa%3A32100/attachment/ATT-0/.
 |
