Biology:Sib RNA
| Sib RNA | |
|---|---|
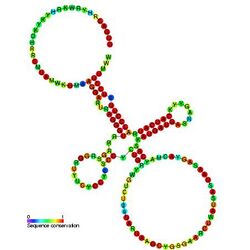 Predicted secondary structure and sequence conservation of QUAD | |
| Identifiers | |
| Symbol | QUAD |
| Alt. Symbols | Sib RNA, QUAD RNA |
| Rfam | RF00113 |
| Other data | |
| RNA type | Gene; sRNA |
| Domain(s) | Bacteria |
| SO | 0000655 |
| PDB structures | PDBe |
| Ibs Toxin of Type I toxin-antitoxin system | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Ibs_toxin | ||||||||
| Pfam | PF13957 | ||||||||
| Membranome | 391 | ||||||||
| |||||||||
Sib RNA refers to a group of related non-coding RNA. They were originally named QUAD RNA after they were discovered as four repeat elements in Escherichia coli intergenic regions.[1] The family was later renamed Sib (for short intergenic abundant sequences) when it was discovered that the number of repeats is variable in other species and in other E. coli strains.[2]
Identification
These small RNA were identified computationally by searching the genome of E. coli for intergenic regions of high sequence identity (sequence conservation) with the genomes of closely related bacteria (several salmonella species and Klebsiella pneumoniae).[3] This data was combined with microarray expression analysis and potential novel ncRNAs identified. The expression of novel ncRNA of interest was confirmed by northern blotting.
In this large scale screen these ncRNAs were simply referred to as candidates 43, 55 and 61.[3] These 3 ncRNA appear to be highly homologous and are derived from a repeat region of the genome. Each of the ncRNA contains a short stretch homologous to boxC, a repeat element of unknown function present in 50 copies or more within the genome of E. coli.[4]
Function
Sib RNA regulates the expression of a toxic protein in a type I toxin-antitoxin system similar to that of hok/sok andldr-rdl genes.[5] The constitutively expressed Sib transcript regulates the ibs (induction brings stasis) open reading frame which encodes a small 18–19 amino acid hydrophobic protein which slows growth at moderate levels of expression and is toxic when overexpressed. The ibs gene is on the opposite strand to sib and is completely complementary, so the antisense-binding of Sib RNA with the ibs mRNA brings about dsRNA-mediated degradation.[2]
When sib was deleted in multi-copy plasmids, the cells could not be maintained due to the toxicity of the unrepressed ibs protein. The toxicity mechanism of ibs protein is not fully understood, but a change in membrane potential upon over-expression of the protein suggests that interactions with membrane proteins or membrane insertion brings about cell death.[2]
See also
References
- ↑ Rudd KE (1999). "Novel intergenic repeats of Escherichia coli K-12". Res. Microbiol. 150 (9–10): 653–664. doi:10.1016/S0923-2508(99)00126-6. PMID 10673004.
- ↑ 2.0 2.1 2.2 "Repression of small toxic protein synthesis by the Sib and OhsC small RNAs". Mol. Microbiol. 70 (5): 1076–1093. December 2008. doi:10.1111/j.1365-2958.2008.06394.x. PMID 18710431.
- ↑ 3.0 3.1 "Identification of novel small RNAs using comparative genomics and microarrays". Genes Dev. 15 (13): 1637–1651. 2001. doi:10.1101/gad.901001. PMID 11445539.
- ↑ Bachellier, S., Gilson, E., Hofnung, M., and Hill, C.W. 1996. Repeated sequences. In Escherichia coli and Salmonella: Cellular and molecular biology (ed. F.C. Neidhardt, et al.), pp. 2012–2040. American Society for Microbiology, Washington, D.C.
- ↑ "Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli". Mol. Microbiol. 45 (2): 333–349. July 2002. doi:10.1046/j.1365-2958.2002.03042.x. PMID 12123448.
Further reading
- "Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families". Nucleic Acids Res. 38 (11): 3743–3759. June 2010. doi:10.1093/nar/gkq054. PMID 20156992.
- "Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family.". Nucleic Acids Res 38 (17): 5851–5866. 2010. doi:10.1093/nar/gkq292. PMID 20453032.
- "Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110". Mol. Syst. Biol. 2 (1): 2006.0007. 2006. doi:10.1038/msb4100049. PMID 16738553.
- "Regulatory RNA in bacterial pathogens". Cell Host Microbe 8 (1): 116–127. July 2010. doi:10.1016/j.chom.2010.06.008. PMID 20638647.
- Rudd KE (1999). "Novel intergenic repeats of Escherichia coli K-12". Res. Microbiol. 150 (9–10): 653–664. doi:10.1016/S0923-2508(99)00126-6. PMID 10673004.
External links
 |
