Biology:Tilapia tilapinevirus
| Tilapia tilapinevirus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Amnoonviridae |
| Genus: | Tilapinevirus |
| Species: | Tilapia tilapinevirus
|
| Synonyms[1] | |
| |
Tilapia tilapinevirus, or Tilapia lake virus (TiLV), is a negative-strand RNA virus that infects both wild and aquacultured populations of tilapia.[2] It is the only species in the monotypic genus Tilapinevirus, which in turn is the only genus in the family Amnoonviridae.[3] Thus far it has been recorded in various regions across Asia, Africa, and South America.[2] The virus was first discovered and identified in 2014 when the Sea of Galilee (Kinneret Lake) in Israel experienced a major noticeable decline in tilapia catch quantities.[4]
Classification
Tilapia lake virus is a negative-sense, single-stranded RNA virus.[4] It belongs to Group V of the Baltimore Classification System of viruses.[citation needed]
Structure
Electron microscopy has revealed tilapia lake virus to be an enveloped particle with a helical nucleocapsid[4] that is 55–100 nm in diameter.[2] Further information of the viral structure is not yet available, however, as TiLV is described to be an orthomyxo-like virus[5] it may share similar structural features, like surface glycoproteins and a helical nucleocapsid,[6] as other spherical orthomyxoviruses.
Genome
The RNA strand is segmented in ten viral genomic segments with open reading frames (ORF) which encode for ten proteins.[7] The genome's total size is 10.323kb and each of the ten segments range in size from 465 to 1,641 nucleotides.[5] The first, and largest, of the segments has minimal homology with the influenza C virus PB1 subunit. The remaining nine segments show no homology with other known viruses, though their genome organization is consistent with that of other orthomyxoviruses.[4] Comparative genome analysis of TiLV from various global populations of tilapia has indicated that the genome segments have geographically influenced genetic variation.[8] 13 nucleotides that are present in all segments are also included in the TiLV 5' and 3' noncoding termini, which gives TiLV resemblance to two other orthomyxoviruses, Isavirus and Thogoto.[5]
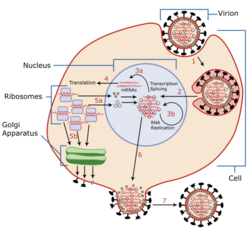
Replication cycle
Attachment and entry
There is limited information of the replication cycle specific to TiLV, but it is known to be in the family Amnoonviridae due to its single-stranded negative-sense segmented RNA genome. Scientists previously thought it might belong to the family Orthomyxoviridae because they carry several surface glycoproteins that recognize and bind to sialic acid receptors on the target cell membrane.[9] The target cell transports the virus into the cell by receptor-mediated endocytosis, initiating endosome acidification.[10] This acidification results in a conformational change of the viral glycoprotein, initiating membrane fusion of the viral envelope and endosomal membrane. Once fusion is complete, the viral genome, accessory proteins, and RNA dependent RNA polymerase are released into the host cell cytoplasm.[10]
Replication and transcription
Through in situ hybridization, it is found that transcription of the TiLV viral genome occurs in the nucleus, this is common among all Orthomyxoviridae.[5] The nucleocapsid of an orthomyxovirus is transported to the nucleus where it is transcribed by viral enzymes resulting in the production of viral mRNA.[11] Cap sequences are taken from the host cell mRNA during transcription and bound to viral mRNA, this allows the viral mRNA to exit the nucleus and return to the cytoplasm of the host cell where it will by recognized and translated into proteins by host cell ribosomes.[11] The 5' and 3' noncoding termini of TiLV include 13 similar nucleotides, this enables base pairing and replication, transcription, and packaging of viral RNA as a result of the formation of secondary structures.[12] In addition, all of the 5′ ends of TiLV genomic RNA segments contain a brief uridine stretch (3 to 5 bases long). This short uninterrupted sequence resembles that of the 5 to 7 uridine nucleotides found in many other orthomyxoviridae, this occurs when the viral polymerase "stutters" while assembling poly(A) tails.[5]
Assembly and release
Orthomyxovirus viral glycoproteins then travel to the cell membrane where they form a spherical bud to transport negative-stranded vRNA out of the host cell. After the new viral material leaves the host cell, the host cell is terminated.[13]
Interactions with host
In cell culture, the affected cells exhibit significant cytopathic effect (CPE), structural changes of the host cell due to viral infection.[4] Clear and rapid CPE development occurs primarily at the E-11 cell line, cell lines of the brain and liver have been shown to be highly permissive at propagating TiLV.[2][4] Cases of infection note syncytium formation, the fusion of infected neighboring cells to produce multi-nucleated cells. Syncytial cells of this species are characterized by swollen mitochondria.[8] Hepatocytes of infected tilapia are swollen and dissociated,[8] with significant cytoplasmic accumulation of yellow to brown pigment (MMC) in both the spleen and liver of naturally and experimentally infected fish.[4] 'in addition, experimental infection shows histologic lesions on the brain such as edema, focal hemorrhages in the leptomeninges, and capillary congestion in both the white and gray matter.[4]
Studies have shown that upon experimental infection of TiLV, histopathological lesions have been found similar to those seen in natural outbreaks.[4] These natural outbreaks have been characterized with lethargy, discoloration, ocular alterations, skin patches, and ulcerations of the digestive tract.[4] The main organs where pathology is observed are the brain, eyes, and liver. Gross lesions are commonly visible in infected species such as ocular opacity of the cataract, and skin erosions such as,[4] loss of scales or discoloration, skin hemorrhages, abdominal swelling, scale protrusion, and exophthalmia.[12] At this time, there is no vaccine for TiLV and it has an >80% mortality rate.[4]
Tropism
TiLV pathology has been found to impact most the optic, brain, and liver tissues.[4] However, some studies suggest that TiLV tropism may also occur in connective muscle tissues, kidneys, gills, spleen, and heart.[8] The tropism of the virus seems to vary based on geographic location. Laboratory studies on tilapia in Israel cited lethargy, ocular alterations and lesions, and skin erosions in both wild and farmed tilapia, with discoloration also as prevalent sign of TiLV among the farmed tilapia.[4] Some Egyptian farms' TiLV infected fish displayed hemorrhagic patches, detached scales, open wounds, discoloration, and fin rot though some were co-infected with aeromonas .[14] Tilapia observed in Ecuador have displayed discoloration as well, along with exophthalmia, abdominal distension, protrusion of the scales, and gill pallor.[15] Exophthalmia and ulcers have accompanied TiLV in Peruvian tilapia.[16] Skin congestion and erosion, discoloration, abnormal behavior, lethargy, loss of appetite, pallor, anemia, exophthalmia, and abdominal swelling have been observed signs of pathology in Thailand.[8][7] In India, naturally infected tilapia had skin erosions and loss of scales as a result of infection.[17] Those infected with TiLV in the laboratory setting in India exhibited exophthalmia, abdominal swelling, and scale protrusion.[17] Reports of TiLV cases in the Philippines cited abdominal swelling and exophthalmia as results of TiLV infection.[18]
Associated diseases
There is little known of the relationship of TiLV to other viral aquaculture diseases, but viruses such as infectious salmon anemia orthomyxoviruses, infectious hematopoietic necrosis virus, and viral hemorrhagic septicemia virus are common causes of illness and death in cultured fish.[7] Salmon anemia orthomyxoviruses, influenza, and Thogoto have also been said to have similar replication to that of TiLV due to organization of nucleotide sequences in transcription enabling base pairing.[5] Specific to tilapia, Tilapia iridovirus is the only significant viral pathogen known to cause severe disease and die-offs.[19] Other pathogens known to cause viral diseases in this species are betanodavirus and herpes-like virus.[7]
Transmission and control
The virus is found to be transmitted through direct horizontal transmission by cohabitation or transfer of live aquatic animals, although these viral pathogens have been found in fresh and preserved tilapia.[20][21] There is limited information on the biophysical properties and risks of TiLV associated with animal product but research suggests that the eye, brain and liver are likely to contain highest concentrations of TiLV and thus solid and liquid waste are likely to be contaminated.[20] There is currently no evidence of vertical transmission of TiLV.[citation needed]
Restriction of the movement of live tilapines between farms or fisheries is thought to limit the spread of the viral disease to new species, as well as maintaining clean practices and sanitizing equipment in these areas. There is still no evidence that there are practices to limit viral spread in an infected farm.[20]
Significance
The spread of TiLV has global impact in both commercial and ecological settings. Worldwide trade of tilapia has huge economic impact as an industry that brings in approximately 4.5 million metric tons of product and $7.5 billion annually.[5] This industry is a major employer in China, Egypt, Thailand, Philippines, Indonesia, Laos, Costa Rica, Colombia, Ecuador and Honduras with the United States as the lead importer.[5] Tilapia are also very important for ecological systems[4] as they are beneficial in algae, mosquito, and general habitat control.[5]
References
- ↑ Bacharach, Eran; Mishra, Nischay; Briese, Thomas; Eldar, Avi; Lipkin, W. Ian; Kuhn, Jens H. (18 July 2016). "Create one (1) new genusincluding one (1) new species" (in en). https://ictv.global/ICTV/proposals/2016.016a-dM.A.v2.Tilapinevirus.pdf.
- ↑ 2.0 2.1 2.2 2.3 "Tilapia lake virus: a threat to the global tilapia industry?". Reviews in Aquaculture 11 (3): 725–739. 2018. doi:10.1111/raq.12254.
- ↑ "Virus Taxonomy: 2018b Release" (in en). February 2019. https://ictv.global/taxonomy.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 "Identification of a novel RNA virus lethal to tilapia". Journal of Clinical Microbiology 52 (12): 4137–46. December 2014. doi:10.1128/JCM.00827-14. PMID 25232154.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 "Characterization of a Novel Orthomyxo-like Virus Causing Mass Die-Offs of Tilapia". mBio 7 (2): e00431-16. April 2016. doi:10.1128/mBio.00431-16. PMID 27048802.
- ↑ "Medical Definition of ORTHOMYXOVIRIDAE" (in en). https://www.merriam-webster.com/medical/Orthomyxoviridae.
- ↑ 7.0 7.1 7.2 7.3 "Outbreaks of Tilapia Lake Virus Infection, Thailand, 2015–2016". Emerging Infectious Diseases 23 (6): 1031–1033. June 2017. doi:10.3201/eid2306.161278. PMID 28518020.
- ↑ 8.0 8.1 8.2 8.3 8.4 "Emergence of tilapia lake virus in Thailand and an alternative semi-nested RT-PCR for detection.". Aquaculture 476: 111–8. July 2017. doi:10.1016/j.aquaculture.2017.04.019.
- ↑ "Functional balance between haemagglutinin and neuraminidase in influenza virus infections". Reviews in Medical Virology 12 (3): 159–66. May 2002. doi:10.1002/rmv.352. PMID 11987141.
- ↑ 10.0 10.1 "Chapter 7 - Viruses as Pathogens: Animal Viruses, With Emphasis on Human Viruses". Viruses: Molecular Biology, Host Interactions and Applications to Biotechnology. Academic Press. 2018. pp. 157–187. doi:10.1016/B978-0-12-811257-1.00007-3. ISBN 9780128112571.
- ↑ 11.0 11.1 Orthomyxoviruses (4th ed.). University of Texas Medical Branch at Galveston. 1996. ISBN 9780963117212. http://www.ncbi.nlm.nih.gov/books/NBK8611/. Retrieved 2019-03-13.
- ↑ 12.0 12.1 Fenwick, Brad, ed (March 2017). "Detection of Tilapia Lake Virus in Clinical Samples by Culturing and Nested Reverse Transcription-PCR". Journal of Clinical Microbiology 55 (3): 759–767. doi:10.1128/JCM.01808-16. PMID 27974544.
- ↑ "Assembly and budding of influenza virus". Virus Research 106 (2): 147–65. December 2004. doi:10.1016/j.virusres.2004.08.012. PMID 15567494.
- ↑ "Detection of Tilapia Lake Virus in Egyptian fish farms experiencing high mortalities in 2015". Journal of Fish Diseases 40 (12): 1925–1928. December 2017. doi:10.1111/jfd.12650. PMID 28590067.
- ↑ "Syncytial hepatitis of farmed tilapia, Oreochromis niloticus (L.): a case report". Journal of Fish Diseases 37 (6): 583–9. June 2014. doi:10.1111/jfd.12142. PMID 23802941.
- ↑ "Tilapia lake virus, Peru". http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=26027.
- ↑ 17.0 17.1 "Emergence of tilapia lake virus associated with mortalities of farmed Nile tilapia Oreochromis niloticus (Linnaeus 1758) in India.". Aquaculture 484: 168–74. February 2018. doi:10.1016/j.aquaculture.2017.11.025.
- ↑ "Tilapia Lake Virus (TiLV), Philippines". http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=25278.
- ↑ "Tilapia Diseases" (in en-US). http://americulture.com/tilapia-diseases.
- ↑ 20.0 20.1 20.2 "Tilapia lake virus (TiLV)—a novel orthomyxo-like virus". World Organisation for Animal Health (OIE). February 2018. http://www.oie.int/fileadmin/Home/eng/Internationa_Standard_Setting/docs/pdf/A_TiLV_disease_card.pdf.
- ↑ Fenwick, Brad, ed (March 2017). "Detection of Tilapia Lake Virus in Clinical Samples by Culturing and Nested Reverse Transcription-PCR". Journal of Clinical Microbiology 55 (3): 759–767. doi:10.1128/JCM.01808-16. PMID 27974544.
Wikidata ☰ Q23758055 entry
 |