Biology:Uncharacterized LOC644249 gene
Uncharacterized LOC644249 gene.,[1] also known as RP11-195B21.3, is about 1058 base pairs long and is found in Homo sapiens on chromosome 9q12. More specifically, the sequence is located on Chromosome: 9; NC_000009.11(67977457..67987991 bp). This gene’s protein product is the “coiled-coil domain-containing protein 29” which is 291 amino acids long and may contain a conserved domain in the superfamily, pfam 12001. In particular, this conserved domain contains the domain of unknown function DUF3496 which is about 110 amino acids long, functionally uncharacterized, and found in eukaryotes.[2] Other possible motifs for the protein product exist but the DUF3496 remains the most likely.[3] This protein may play a role as a transmembrane protein.[4]
Gene
Genomic Location
For humans, this gene is located on chromosome 9q12 (67977457..67987991) bp.[1]
Promoter
The promoter is predicted to be about 601 bp in length and is about 100 bp upstream from the transcription start site of the primary transcript .[5]
Transcript
This gene is predicted to have 4 exons, with a very small 3'UTR which is capped with a poly A tail very shortly after the 4th exon.[6]
Protein
The protein product is the coiled-coil domain-containing protein 29, abbreviated CCDC29, is about 291 amino acids in length and contains the DUF3496 motif. Also, this protein is also predicted to have a possible transmembrane site at about 284 - 291 aa.[4]
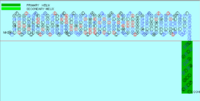
Charge Distribution Analysis
1 0000000000 0--0-000-0 +0-00+0-00 0++0+0+0-- 000-+-0000 +00000+-00
61 0000-0000+ +00000-+0- --++00+--0 000+000-00 +0-00+00-0 +0-0--+000
121 00-+0--000 0-0000-+00 -000+-0000 00++-000+- 0-0+00-0+0 00–000-0-
181 -0+-000000 +000000++0 0+00++0000 00+000-+-0 0+00000000 ++-0-0000-
241 00000000++ 0000000+00 0000000000 +000000000 0000000000 0[4]
Motif
The DUF3496 motif[2] is about 110 aa long and is conserved within eukaryotes. As the name implies, it is currently unknown in function but can be found across different species. On CCDC 29, its relative location is about 153 - 259 aa.
Expression
This GEO profile of LOC644249 was included in a gene expression profile which was conducted in a study[7] that analyzed adipose tissues of subjects at risk of metabolic syndrome. This GEO profile may suggest that LOC644249 is ubiquitously expressed in adipose tissue but no direct correlation of the effects of the expression levels of LOC644249 with adipose tissue can be made.[4]
Transcript Variant
Only two transcript variants were predicted to might occur. One transcript variant predicted the loss of exon 4. The loss of exon 4 would lead to the loss of roughly half of the DUF3496 and the entire loss of the transmembrane domain. The other transcript variant predicted the loss of exon 2, which would not lead to the loss of DUF3496 and leaves the reading frame relatively intact.[4][8]
Homology
Orthologs of CCDC29 seem to be only limited to primates as shown by the table below. A paralog of CCDC29 which is not only limited to primates, Ankyrin repeat domain-containing protein 26, showed that this paralog is reserved within vertebrate animals only. The most distant ortholog of Ankyrin repeat domain-containing protein 26 was found to be the gallus gallus species.
| Scientific Name | Common Name | Accession number | Sequence Length (aa) | Percent Identity | Percent Similarity |
|---|---|---|---|---|---|
| Homo sapiens | Human | XP_003960494.1 | 291 | - | - |
| Gorilla gorilla | Gorilla | XP_004062636.1 | 223 | 86% | 94% |
| Sus scrofa | chimpanzee | XP_003954217.1 | 290 | 89% | 97% |
| Nomascus leucogenys | Northern White-cheeked Gibbon | XP_004087823.1 | 273 | 89% | 97% |
Protein Post-Modification
N glycosylation
One possible N-glycosylation site was predicted, but a signal peptide was not detected. Thus, it is possible that CCDC29 does not undergo this particular modification even though it has the possible site.[9]
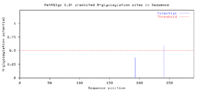
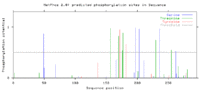
Phosphorylation
A total of 13 likely phosphorylation sites were predicted: Ser: 5 Thr: 6 Tyr: 2
The main concentration of the phosphorylation sites seem to be localized within the DUF3496 motif and the 3' end of the protein.[10]
Protein structure
The CCDC29, as the name implies, is thought to have the coiled coil motif. Protein structure prediction software confirms this motif 97.6% confidence.[11]
Transmembrane
CCDC29 is predicted to have one transmembrane helix located at the N terminal 270.[12]
Function
Currently unknown as of 5/9/2013.
References
- ↑ 1.0 1.1 "NCBI Gene: LOC644249 summary". https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=644249.
- ↑ 2.0 2.1 "NCBI PFAM". https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?seqinput=XP_003960494.1.
- ↑ "Wellcome trust Sanger Institute". http://pfam.sanger.ac.uk/search/sequence.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 "SDSC Biology WorkBench". http://workbench.sdsc.edu/.
- ↑ "Genomatix Genome Analysis Tool". http://www.genomatix.de/.
- ↑ "Softberry FGENESH". http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind.
- ↑ Van Dijk, Susan J.; Feskens, Edith JM; Bos, Marieke B.; Hoelen, Dianne WM; Heijligenberg, Rik; Bromhaar, Mechteld Grootte; De Groot, Lisette Cpgm; De Vries, Jeanne HM et al. (Dec 2009). "A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome". Am J Clin Nutr 90 (6): 1656–64. doi:10.3945/ajcn.2009.27792. PMID 19828712.
- ↑ "UCSC Genome Bioinformatics". http://genome.ucsc.edu/index.html.
- ↑ "ExPASy N-glycosylation". http://www.cbs.dtu.dk/services/NetNGlyc/.
- ↑ "ExPASy Phosphorylation". http://www.cbs.dtu.dk/services/NetPhos/.
- ↑ "ExPASy Phyre2". http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index.
- ↑ "ExPASy SOSUI". http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html.
 |