Chemistry:Desosamine
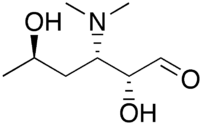
| |
| Names | |
|---|---|
| IUPAC name
3,4,6-Trideoxy-3-(dimethylamino)-D-xylo-hexose
| |
| Systematic IUPAC name
(2R,3S,5R)-3-(Dimethylamino)-2,5-dihydroxyhexanal | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C8H17NO3 | |
| Molar mass | 175.23 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Desosamine is a 3-(dimethylamino)-3,4,6-trideoxyhexose found in certain macrolide antibiotics (contain a high level of microbial resistance) such as the commonly prescribed erythromycin,[1][2] azithromycin, clarithroymcin, methymycin, narbomycin, oleandomycin, picromycin and roxithromycin. As the name suggests, these macrolide antibiotics contain a macrolide or lactone ring and they are attached to the ring Desosamine which is crucial for bactericidal activity.[3] The biological action of the desosamine-based macrolide antibiotics is to inhibit the bacterial ribosomal protein synthesis.[4] These antibiotics which contain Desosamine are widely used to cure bacterial-causing infections in human respiratory system, skin, muscle tissues, and urethra.
Discovery
Although desosamine has been found in many macrolide antibiotics, the complete chemical structure of desosamine was not determined until 1962.[5] Nuclear magnetic resonance spectroscopy data was used to establish the complete configuration of desosamine. The hydrogen atoms at the C1,C2,C3, and C5 positions are all found to be axial.[5]
Biosynthesis
Six enzymes are required for Desosamine biosynthesis from TDP-glucose in Streptomyces venezuelae.[1][6] In addition to the required enzymes, there are eight important open reading frames known as the des regions, they are desI~desVIII, these eight frames are the necessary genes used in Desosamine biosynthesis, among the 8 des regions, the desI gene implements C-4 Deoxygenation by the enzymatic activity of dehydrase. [7]
Degradation
Degradation of several of the aforementioned antibiotics yields the desosamine sugar. It is found in combination with the smaller macrolide rings, always attached at C-3 or C-5 of the aglycone. Alkaline degradation found the sugar to be a D-hexose derivative.[8] Glycosidic cleavage of methomycin produces aglycone methynolide and the basic sugar desosamine, whose structure had been determined by oxidative degradation to crotonaldehyde and by other experiments.[9]
Drug resistance
Macrolide antibiotics that contain Desosamine as an amino sugar in their chemical structures sometimes encounter drug-resistant bacteria. The target-site modification can result in changing chemical structure of the antibiotics, for example, a methylation mutation, which will block the drug from normally functioning.[10]
See also
References
- ↑ 1.0 1.1 "In vivo characterization of the dTDP-D-desosamine pathway of the megalomicin gene cluster from Micromonospora megalomicea". Microbiology 152 (Pt 3): 667–673. March 2006. doi:10.1099/mic.0.28680-0. PMID 16514147.
- ↑ "Microbial degradation of erythromycins A and B". The Journal of Antibiotics 28 (4): 307–11. April 1975. doi:10.7164/antibiotics.28.307. PMID 1150530.
- ↑ "Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine". Biochemistry 46 (31): 8999–9006. August 2007. doi:10.1021/bi700751d. PMID 17630700.
- ↑ "How Macrolide Antibiotics Work". Trends in Biochemical Sciences 43 (9): 668–684. September 2018. doi:10.1016/j.tibs.2018.06.011. PMID 30054232.
- ↑ 5.0 5.1 "The stereochemistry of desosamine, an nmr analysis" (in en). Tetrahedron Letters 3 (17): 735–739. January 1962. doi:10.1016/S0040-4039(00)70510-7.
- ↑ "Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine". Biochemistry 46 (31): 8999–9006. August 2007. doi:10.1021/bi700751d. PMID 17630700.
- ↑ "Biosynthesis of desosamine: construction of a new macrolide carrying a genetically designed sugar moiety". Organic Letters 1 (1): 133–6. July 1999. doi:10.1021/ol9906007. PMID 10822548.
- ↑ "Carbohydrate components of antibiotics. Part I. Degradation of desosamine by alkali: Its absolute configuration at position 5". Journal of the Chemical Society 1961 (4): 4831–4836. 1961. doi:10.1039/JR9610004831.
- ↑ "The macrolide antibiotics". Quarterly Reviews, Chemical Society 17 (4): 343–361. 1963. doi:10.1039/QR9631700343.
- ↑ "Hidden epidemic of macrolide-resistant pneumococci" (in en-us). Emerging Infectious Diseases 11 (6): 802–7. June 2005. doi:10.3201/eid1106.050147. PMID 15963272.
External links
- Desosamine in the US National Library of Medicine MeSH (Medical Subject Headings):
- Deoxy Sugar in the US National Library of Medicine MeSH (Medical Subject Headings)
 |
