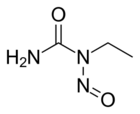
Chemistry:ENU
| |
| |
| Names | |
|---|---|
| Preferred IUPAC name
N-Ethyl-N-nitrosourea | |
Other names
| |
| Identifiers | |
3D model (JSmol)
|
|
| Abbreviations | ENU[citation needed] |
| 1761174 | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| EC Number |
|
| KEGG | |
PubChem CID
|
|
| RTECS number |
|
| UNII | |
| UN number | 2811 |
| |
| |
| Properties | |
| C3H7N3O2 | |
| Molar mass | 117.108 g·mol−1 |
| log P | 0.208 |
| Vapor pressure | 0.00244 kPa @ 25˚C[1] |
| Acidity (pKa) | 12.317 |
| Basicity (pKb) | 1.680 |
| Absorbance | ε398 = 11.86 mM−1 cm−1[2] |
| Hazards | |
| GHS pictograms |  
|
| GHS Signal word | DANGER |
| H301, H312, H332, H350, H360 | |
| P280, P308+313 | |
| Lethal dose or concentration (LD, LC): | |
LD50 (median dose)
|
300 mg kg−1 (oral, rat) |
| Related compounds | |
Related ureas
|
|
Related compounds
|
|
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
ENU, also known as N-ethyl-N-nitrosourea (chemical formula C3H7N3O2), is a highly potent mutagen. For a given gene in mice, ENU can induce 1 new mutation in every 700 loci. It is also toxic at high doses.
The chemical is an alkylating agent, and acts by transferring the ethyl group of ENU to nucleobases (usually thymine) in nucleic acids. Its main targets are the spermatogonial stem cells, from which mature sperm are derived.
Background of discovery of ENU as a mutagen
Bill Russell (1951) created a landmark in the field of mouse genetics by creating a specifically designed mouse strain, the T (test) stock that was used in genetic screens for testing mutagens such as radiations and chemicals. The T-stock mouse harbors 7 recessive, viable mutations affecting easily recognizable traits. At the Oak Ridge National Laboratory, Russell's initial goal was to determine the rate of inheritable gene mutations in the germ line induced by radiations. Thus he decided to use T-stock mice in order to define how often a set of loci could be mutated with radiations. Since the mutations in the T-stock mouse were recessive, the progeny would have a wild type phenotype (as a result of crossing a mutant [e.g.s/s mutant male] to a wild type female [+/+]). Thus with any progeny carrying a mutation induced by radiation at one of the 7 loci, would exhibit the mutant phenotype in the first generation itself. This approach, the specific locus test (SLT) allowed Russell to study a wide range of specific mutations and to calculate the mutation rates induced by radiations.[3]
In addition to studying the effect of radiation for SLT, Russell et al. were also interested in studying the effect of chemical mutagens such as procarbazine and ethylnitrosourea for SLT. At that time, procarbazine was the most potent chemical mutagen known to cause a significant spermatogonial mutagenesis in an SLT, although at a rate one-third of that of X-rays. Russell's earlier mutagenesis work on Drosophila using diethylnitrosoamine (DEN) triggered them to use DEN for the SLT. However, DEN needs to be enzymatically converted into an alkylating agent in order to be mutagenic and probably this enzymatic activation was not sufficient in mammals. This could be illustrated by the extremely low mutation rate in mice given by DEN (3 in 60,179 offspring). To overcome this problem, a new mutagen, N-ethyl N-nitrosourea (ENU), an alkylating agent, which does not need to be metabolised, was suggested to be used by Ekkehart Vegel to Russell et al. The ENU (250 mg/kg) induced mice underwent a period of sterility for 10 weeks. After recovery, 90 males were crossed to the T-stock females and 7584 pups were obtained.[3] Their results showed that a dose of 250 mg/kg of ENU was capable of producing a mutation rate 5 times higher than that obtained with 600R (1R = 2.6 x10^-4 coulombs/kg) of acute X-irradiation. This rate was also 15 times higher to that obtained with procarbazine (600 mg/kg).[4]
To overcome the problem of initial period of sterility, the Russell group showed that instead of injecting one large dose of ENU, a fractionated dose (100 mg/kg)[5] on a weekly schedule permitted a total higher dose (300–400 mg/kg)[5] to be tolerated. This further showed that the mutation frequency improved to be 12 times that of X-rays, 36 times that of procarbazine and over 200 times that of spontaneous mutations. When the mutation rate was averaged across all 7 loci, ENU was found to induce mutations at a frequency of one per locus in every 700 gametes.[3]
Summary of properties and advantages of ENU mutagenesis
- ENU is an alkylating agent and has preference for A->T base transversions and also for AT->GC transitions.[6] However it is also shown to cause GC->AT transitions.[7]
- It is known to induce point mutations, which implies that by mapping for the desired phenotype, the researcher can identify a single candidate gene responsible for the phenotype.[8]
- The point mutations are at approximately 1-2 Mb (Mega-base-pair) interval and occur at an approximate rate of 1 per 700 gametes.[3]
- ENU targets spermatogonial stem cells.[6]
ENU - A genetic tool in mutagenesis screens: Overview
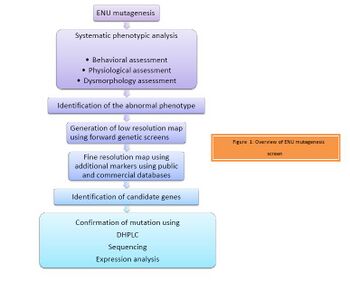
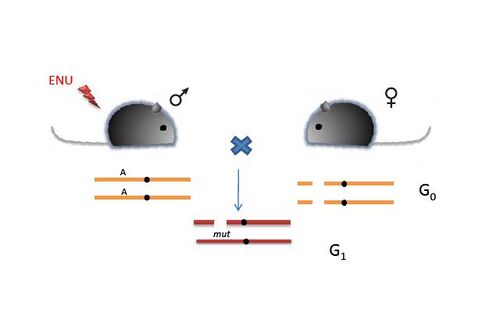
Ever since the discovery of ENU as the most potent mutagen by Russell et al. it has been used in forward (phenotype based) genetic screens with which one can identify and study a phenotype of interest. As illustrated in Figure 1, the screening process begins with mutagenising a male mouse with ENU. This is followed by systematic phenotypic analysis of the progeny. The progeny are assessed for behavioral, physiological or dysmorphological changes. The abnormal phenotype is identified. Identification of the candidate gene is then achieved by positional cloning of the mutant mice with the phenotype of interest.
Types of screens
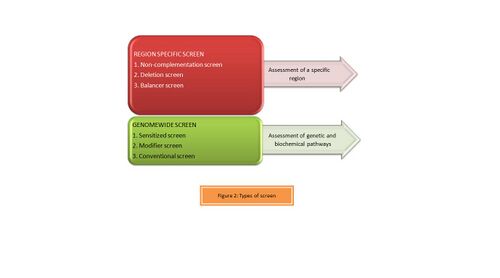
ENU is used as a genetic tool by designing a variety of genetic screens suitable to the researchers' interests. Depending on the region being assessed, forward genetic screens can be classified as illustrated in Figure 2 as:[8]
- Region Specific screens: Studies are designed specifically to obtain a gradient of phenotypes by generating an allelic series which are helpful in studying the region of interest.
- Genome-wide screens: These are simple dominant or recessive screens and are often useful in understanding specific genetic and biochemical pathways.
Region-specific screens
Region specific can be classified as follows:

Non-complementation screens
Complementation is the phenomenon which enables generation of the wild type phenotype when organisms carrying recessive mutations in different genes are crossed.[8] Thus if an organism has one functional copy of the gene, then this functional copy is capable of complementing the mutated or the lost copy of the gene. In contrast, if both the copies of the gene are mutated or lost, then this will lead to allelic non-complementation (Figure 3) and thus manifestation of the phenotype.
The phenomenon of redundancy explains that often multiple genes are able to compensate for the loss of a particular gene. However, if two or more genes involved in the same biological processes or pathways are lost, then this leads to non-allelic non-complementation. In a non-complementation screen, an ENU-induced male is crossed with a female carrying a mutant allele (a) of the gene of interest (A). If the mutation is dominant, then it will be present in every generation. However, if the mutation is recessive or if the G1 progeny are non-viable, then a different strategy is used to identify the mutation. An ENU-treated male is crossed with a wild type female. From the pool of G1 individuals, a heterozygous male is crossed to a female carrying the mutant allele (a). If the G2 progeny are infertile or non-viable, they can be recovered again from the G1 male.
Deletion screens
Deletions on chromosomes can be spontaneous or induced. In this screen, ENU-treated males are crossed to females homozygous for a deletion of the region of interest. The G1 progeny are compound heterozygotes for the ENU-induced mutation (Figure 4). Also, they are haploid with respect to the genes in the deleted region and thus loss-of-function or gain-of-function due to the ENU-induced mutation is expressed dominantly. Thus deletion screens have an advantage over other recessive screens due to the identification of the mutation in the G1 progeny itself.
Rinchik et al. performed a deletion screen and complementation analysis and were able to isolate 11 independent recessive loci, which were grouped into seven complementation groups on chromosome 7, a region surrounding the albino (Tyr) gene and the pink-eyed dilution (p) gene.[8]
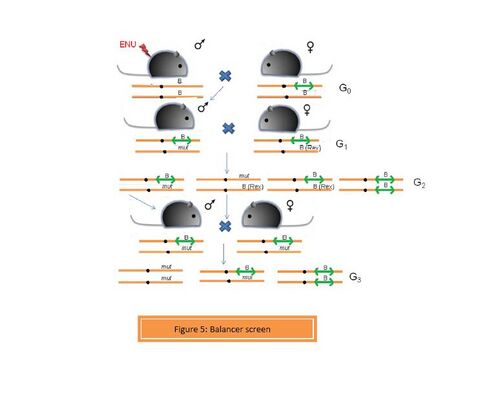
- c. Balancer screens
A chromosome carrying a balancer region is termed as a balancer chromosome. A balancer is a region which prevents recombination between homologous chromosomes during meiosis. This is possible due to the presence of an inverted region or a series of inversions. Balancer chromosome was primarily used for studies in Drosophila melanogaster genetics. Monica Justice et al. (2009) efficiently carried out a balancer screen using a balancer chromosome constructed by Allan Bradley et al. on mouse chromosome 11. In this screen, an ENU-induced male is crossed with a female heterozygous for the balancer chromosome.[8] The mice carrying the balancer chromosome have yellow ears and tail. The G1 heterozygotes are (Figure 5) are crossed to females carrying the rex mutation (Rex in figure 5), which confers a curly coat. In G2, homozygotes for the balancer are non-viable and are not recovered. Mice carrying the rex mutation trans to the balancer or ENU-induced mutation have a curly coat and are discarded. The ENU mutant + rex mutant mice are discarded in order to prevent recombination between those two chromosomes during the next breeding step, which is generating homozygous mutants. Mice that are compound heterozygotes for the balancer and the ENU-induced mutation are brother-sister mated to obtain homozygotes for the ENU-induced mutation in G3.
Genome-wide screens
Genome-wide screens are most often useful for studying genetic diseases in which multiple genetic and biochemical pathways may be involved. Thus with this approach, candidate genes or regions across the genome, that are associated with the phenotype can be identified.
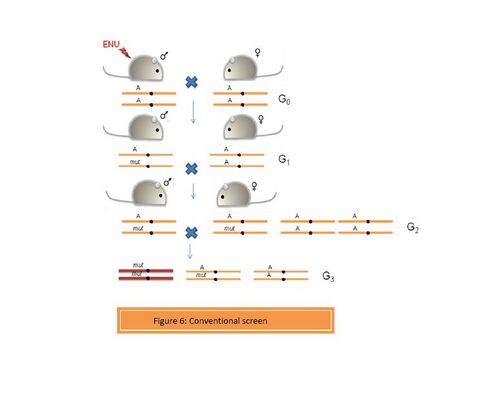
- a. Conventional screens
These screens can be designed to identify simple dominant and recessive phenotypes. (Figure 6). Thus an ENU-induced G0 male is crossed with a wild type female. The G1 progeny can be screened to identify dominant mutations. However, if the mutation is recessive, then G3 individuals homozygous for the mutation can be recovered from the G1 males in two ways:
- A] The G1 male is crossed with a wild type female to generate a pool of G2 progeny. The G3 individuals can be obtained by crossing the G1 male to the G2 daughters. This will yield a proportion of the G3 individuals which resemble the G1 male to a large extent.
- B] G1 male is crossed to a wild type female to obtain a pool of G2 animals., which are then brother-sister mated to obtain the G3 progenies. This approach yields a variety of mutants in the G3 progeny.
A number of organizations around the world are performing genome-wide mutagenesis screens using ENU. Some of them include the Institute of Experimental Genetics at the German Research Center for Environmental Health (GSF), Munich, Germany; The Jackson Laboratory, Maine, USA; the Australian Phenomics Facility at the Australian National University, Canberra, Australia; the Department of Neurobiology and Physiology at Northwestern University, Illinois, USA; the Oak Ridge National Laboratory, Tennessee, USA; the Medical Research Council (MRC) Harwell, Oxfordshire, United Kingdom; the Department of Genetics at The Scripps Research Institute, California, USA; the Mouse Mutagenesis Center for Developmental Defects at Baylor College of Medicine, Texas, USA; and others.[6]
- b. Modifier screens
A modifier such as an enhancer or suppressor can alter the function of a gene. In a modifier screen, an organism with a pre-existing phenotype is selected. Thus, any mutations caused by the mutagen (ENU) can be assessed for their enhancive or suppressive activity.[8] Screening for dominant and recessive mutations is performed in a way similar to the conventional genome-wide screen (Figure 7). A number of modifier screens have been performed on Drosophila. Recently, Aliga et al. performed a dominant modifier screen using ENU-induced mice to identify modifiers of the Notch signaling pathway.[9] Delta 1 is a ligand for the Notch receptor. A homozygous loss-of-function of Delta 1 (Dll1lacZ/lacZ) is embryonically lethal. ENU-treated mice were crossed to Dll1lacZ heterozygotes. 35 mutant lines were generated in G1 of which 7 revealed modifiers of the Notch signaling pathway.
Sensitized screens
In the case of genetic diseases involving multiple genes, mutations in multiple genes contributes to the progression of a disease. Mutation in just one of these genes however, might not contribute significantly to any phenotype. Such "predisposing genes" can be identified using sensitized screens.[10] In this type of a screen, the genetic or environmental background is modified so as to sensitize the mouse to these changes. The idea is that the predisposing genes can be unraveled on a modified genetic or environmental background. Rinchik et al. performed a sensitized screen of mouse mutants predisposed to Diabetic nephropathy. Mice were treated with ENU on a sensitized background of type-1 diabetes. These diabetic mice had a dominant Akita mutation in the insulin-2 gene (Ins2Akita). These mice developed albuminuria, a phenotype that was not observed in the non-diabetic offsprings.[11]
Stability
Generally speaking, ENU is fairly unstable, which makes it easier to inactivate when used as an experimental mutagen, compared to moderately more stable mutagens like EMS. Pure crystalline ENU is sensitive to light and moisture, so should be stored at in cold and dry conditions, and freshly prepared into solutions when needed.[1] In aqueous solutions, ENU rapidly degrades at a basic pH, and protocols call for inactivation of ENU solutions with an equal volume of 0.1M KOH for 24 hours, with or without ambient light exposure to supplement inactivation.[2]
See also
References
- ↑ 1.0 1.1 1.2 "N-Nitroso-N-ethylurea". NIEHS. https://ntp.niehs.nih.gov/ntp/roc/content/profiles/nitrosamines.pdf.
- ↑ 2.0 2.1 Salinger, Andrew P.; Justice, Monica J. (2008). "Mouse Mutagenesis Using N-Ethyl-N-Nitrosourea (ENU): Figure 1.". Cold Spring Harbor Protocols (Cold Spring Harbor Laboratory) 2008 (4): pdb.prot4985. doi:10.1101/pdb.prot4985. ISSN 1940-3402. PMID 21356809.
- ↑ 3.0 3.1 3.2 3.3 Davis, A.P., Justice M.J. An Oak Ridge Legacy: The specific locus test and it role in mouse mutagenesis.Genetics 148,7-12 (1998)
- ↑ Russell W.L., Kelly E.M., Hunsicker P.R., Bangham J.W., Maddux S.C., Phipps E.L. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in mouse. Proc. Natl. Acad. Sci.USA 11, 5818-5819 (1979)
- ↑ 5.0 5.1 Hitotsumachi S., Carpenter D.A., Russell W.L. Dose-Repetition Increases the Mutagenic Effectiveness of N-ethyl-N-nitrosourea in Mouse Spermatogonia. Proc. Natl. Acad. Sci.USA 82, 6619-6621 (1985)
- ↑ 6.0 6.1 6.2 Nolan, P, Hugill, A & Cox, RD,2002, p.278-89
- ↑ Coghill, EL et al.,2002, p.255-6
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Kile, BT & Hilton, DJ 2005, p.557-67
- ↑ Rubio-Aliaga, I. et.al. A genetic screen for modifiers of the delta1-dependent notch signaling function in the mouse. Genetics 175, 1451-1463 (2007)
- ↑ Cordes, S.P. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol Mol Biol Rev 69, 426-439 (2005).
- ↑ Tchekneva, E.E. et al. A sensitized screen of N-ethyl-N-nitrosourea-mutagenized mice identifies dominant mutants predisposed to diabetic nephropathy. J Am Soc Nephrol 18, 103-112 (2007).
External links
- Institute of Experimental Genetics, German Research Center for Environmental Health (GSF), Munich, Germany[yes|permanent dead link|dead link}}]
- The Reproductive Genomics Program, The Jackson Laboratory, Maine, USA
- The Neuroscience Mutagenesis Facility, The Jackson Laboratory, Maine, USA
- The Mouse Heart, Lung, Blood, and Sleep Disorders (HLBS) Center, The Jackson Laboratory, Maine, USA
- Australian Phenomics Facility at the Australian National University, Canberra, Australia
- Mouse Chemical Mutagenesis Core Facility, Department of Neurobiology and Physiology at Northwestern University, Illinois, USA
- Oak Ridge National Laboratory, Tennessee, USA
- Medical Research Council (MRC) Harwell, Oxfordshire, United Kingdom
- Mutagenetix, Department of Genetics, The Scripps Research Institute, California, USA
- Mouse Mutagenesis Center for Developmental Defects, Baylor College of Medicine, Texas, USA
- RIKEN Genomics Science Center, Yokohama Institute, Japan
- PROTOCOL: Mouse Mutagenesis Using N-Ethyl-N-Nitrosourea (ENU)
 |