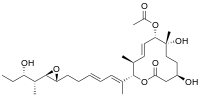
Chemistry:Pladienolide B
| |
| Names | |
|---|---|
| IUPAC name
[(2S,3S,4E,6S,7R,10R)-7,10-dihydroxy-2-[(2E,4E,6S)-7-[(2R,3R)-3-[(2R,3S)-3-hydroxypentan-2-yl]oxiran-2-yl]-6-methylhepta-2,4-dien-2-yl]-3,7-dimethyl-12-oxo-1-oxacyclododec-4-en-6-yl] acetate
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C30H48O8 | |
| Molar mass | 536.706 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Pladienolide B is a polyketide natural product that is made by the bacterium Streptomyces platensis MER-11107,[1][2] which is a gram-positive bacteria isolated from soil in Japan.[3] Pladienolide B is a molecule of interest due to potential anti-cancer properties. The proposed mechanism of action of this molecule is via binding to and inhibiting the SF3B complex within the human spliceosome.[1]
Biosynthesis
As a polyketide, pladienolide B is synthesized in the polyketide synthase (PKS) type I pathway, a biosynthetic pathway that is derived from the fatty acid synthase (FAS) pathway. Polyketides are formed through the condensation of acyl-thioester units. The synthesis of pladienolide B begins with acyl transferase loading propionyl CoA onto the holo (activated) acyl carrier protein (ACP). The molecule is then extended ten times, each time by two carbon units, by iterative PKS (Figure 1).[2] Figure 1 indicates the loading unit (malonyl CoA or methylmalonyl CoA) and tailoring enzymes for each module (turquoise enzyme clusters in part c of Figure 1). After the ten elongation modules, the active site serine residue of thioesterase (TE) acts as a nucleophile by attacking the ketone directly bound to CoA-SH. This nucleophilic attack causes CoA-SH to leave and thus transfers the elongated chain to TE. Then the hydroxyl group on carbon 11 acts as a nucleophile by attacking the terminal ketone so that carbons 1-11 form a cyclic, 12-membered ring, releasing TE in the process. This concludes the PKS portion of the biosynthesis, and thus results in the pladienolide B backbone structure.[2][4]
Post-PKS modifications: Acetyl-transferase (AT) transfers an acetyl group to carbon 7. The following step is either that a P450 I enzyme (PldB C6-hydroxylase) adds a hydroxyl group on carbon 6, and/or that an epoxide group is added between carbons 18 and 19 by P450 II (PldD C18-C19-epoxidase). Based on the purification results of these post-PKS compounds by Boothe et al.,[2] it seems that either hydroxylation or acetylation could occur first, but Boothe et al.[2] found a that hydroxylation seems to occur first more than twice as often as acetylation occurs first.[2] However, regardless of hydroxylation or acetylation occurring first, the result of these tailoring steps yields pladienolide B (Figure 1).

Carbons are numbered using the traditional IUPAC naming system in which C1 is the ketone carbon in the ring, and C23 is the terminal carbon on the chain.[5]
References
- ↑ 1.0 1.1 Trieger, Kelsey A.; La Clair, James J.; Burkart, Michael D. (2020). "Splice Modulation Synergizes Cell Cycle Inhibition". ACS Chemical Biology 15 (3): 669–674. doi:10.1021/acschembio.9b00833. PMID 32004428.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Booth, Thomas J.; Kalaitzis, John A.; Vuong, Daniel; Crombie, Andrew; Lacey, Ernest; Piggott, Andrew M.; Wilkinson, Barrie (2020-08-12). "Production of novel pladienolide analogues through native expression of a pathway-specific activator" (in en). Chemical Science 11 (31): 8249–8255. doi:10.1039/D0SC01928C. ISSN 2041-6539. PMID 34094178.
- ↑ Sakai, Takashi; Sameshima, Tomohiro; Matsufuji, Motoko; Kawamura, Naoto; Dobashi, Kazuyuki; Mizui, Yoshiharu (2004-03-25). "Pladienolides, New Substances from Culture of Streptomyces platensis Mer-11107 I. Taxonomy, Fermentation, Isolation and Screening" (in en). The Journal of Antibiotics 57 (3): 173–179. doi:10.7164/antibiotics.57.173. ISSN 0021-8820. https://www.jstage.jst.go.jp/article/antibiotics1968/57/3/57_3_173/_article.
- ↑ 4.0 4.1 Machida, Kazuhiro; Arisawa, Akira; Takeda, Susumu; Tsuchida, Toshio; Aritoku, Yasuhide; Yoshida, Masashi; Ikeda, Haruo (23 November 2008). "Organization of the Biosynthetic Gene Cluster for the Polyketide Antitumor Macrolide, Pladienolide, in Streptomyces platensis Mer-11107". Bioscience, Biotechnology, and Biochemistry 72 (11): 2946–2952. doi:10.1271/bbb.80425. PMID 18997414.
- ↑ "IUPAC Rules". http://www.chem.uiuc.edu/GenChemReferences/nomenclature_rules.html.
 |
