Neyman Type A distribution
|
Probability mass function 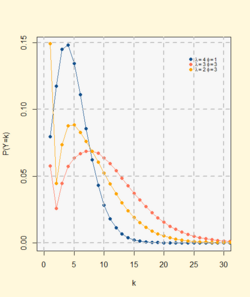 The horizontal axis is the index x, the number of occurrences. The vertical axis we have the probability that the v.a. take the value of x | |||
|
Cumulative distribution function 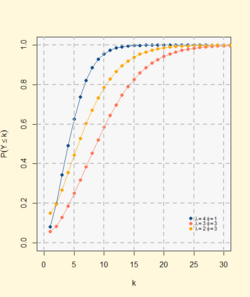 The horizontal axis is the index x, the number of occurrences. The vertical axis we have the acumulative sum of probabilityes from [math]\displaystyle{ P(Y = 0) }[/math] to [math]\displaystyle{ P(Y = x) }[/math] | |||
| Notation | [math]\displaystyle{ \operatorname{NA}(\lambda,\phi) }[/math] | ||
|---|---|---|---|
| Parameters | [math]\displaystyle{ \lambda \gt 0 , \phi \gt 0 }[/math] | ||
| Support | x ∈ { 0, 1, 2, ... } | ||
| pmf | [math]\displaystyle{ \frac{e^{-\lambda}\phi^x}{x!} \sum_{j=0}^{\infty} \frac{(\lambda e^{-\phi})^j j^x}{j!} }[/math] | ||
| CDF | [math]\displaystyle{ e^{-\lambda} \sum_{i=0}^{x}\sum_{j=0}^{\infty} \frac{\phi^i(\lambda e^{-\phi})^j j^i}{i!j!} }[/math] | ||
| Mean | [math]\displaystyle{ \lambda \phi }[/math] | ||
| Variance | [math]\displaystyle{ \lambda \phi(1 + \phi) }[/math] | ||
| Skewness | [math]\displaystyle{ \frac{\lambda \phi(1 + 3\phi + \phi^2)}{(\lambda \phi(1 + \phi))^{3/2}} }[/math] | ||
| Kurtosis | [math]\displaystyle{ \frac{1+7\phi+6\phi^2+\phi^3}{\phi\lambda (1+\phi)^2} }[/math] | ||
| MGF | [math]\displaystyle{ \exp(\lambda(e^{\phi (e^t-1)}-1)) }[/math] | ||
| CF | [math]\displaystyle{ \exp(\lambda(e^{\phi (e^{it}-1)}-1)) }[/math] | ||
| PGF | [math]\displaystyle{ \exp(\lambda(e^{\phi (s-1)}-1)) }[/math] | ||
In statistics and probability, the Neyman Type A distribution is a discrete probability distribution from the family of Compound Poisson distribution. First of all, to easily understand this distribution we will demonstrate it with the following example explained in Univariate Discret Distributions;[1] we have a statistical model of the distribution of larvae in a unit area of field (in a unit of habitat) by assuming that the variation in the number of clusters of eggs per unit area (per unit of habitat) could be represented by a Poisson distribution with parameter [math]\displaystyle{ \lambda }[/math], while the number of larvae developing per cluster of eggs are assumed to have independent Poisson distribution all with the same parameter [math]\displaystyle{ \phi }[/math]. If we want to know how many larvae there are, we define a random variable Y as the sum of the number of larvae hatched in each group (given j groups). Therefore, Y = X1 + X2 + ... X j, where X1,...,Xj are independent Poisson variables with parameter [math]\displaystyle{ \lambda }[/math] and [math]\displaystyle{ \phi }[/math].
History
Jerzy Neyman was born in Russia in April 16 of 1894, he was a Polish statistician who spent the first part of his career in Europe. In 1939 he developed the Neyman Type A distribution [1] to describe the distribution of larvae in experimental field plots. Above all, it is used to describe populations based on contagion, e.g., entomology (Beall[1940],[2] Evans[1953][3]), accidents (Creswell i Froggatt [1963]),[4] and bacteriology.
The original derivation of this distribution was on the basis of a biological model and, presumably, it was expected that a good fit to the data would justify the hypothesized model. However, it is now known that it is possible to derive this distribution from different models (William Feller[1943]),[5] and in view of this, Neyman's distribution derive as Compound Poisson distribution. This interpretation makes them suitable for modelling heterogeneous populations and renders them examples of apparent contagion.
Despite this, the difficulties in dealing with Neyman's Type A arise from the fact that its expressions for probabilities are highly complex. Even estimations of parameters through efficient methods, such as maximum likelihood, are tedious and not easy to understand equations.
Definition
Probability generating function
The probability generating function (pgf) G1(z), which creates N independent Xj random variables, is used to a branching process. Each Xj produces a random number of individuals, where X1, X2,... have the same distribution as X, which is that of X with pgf G2(z). The total number of individuals is then the random variable,[1]
- [math]\displaystyle{ Y = SN = X_1 + X_2 + ... + X_N }[/math]
The p.g.f. of the distribution of SN is :
- [math]\displaystyle{ E[z^{SN}] = E_N[E[z^{SN}|N]] = E_N[G_2(z)] = G_1(G_2(z)) }[/math]
One of the notations, which is particularly helpful, allows us to use a symbolic representation to refer to an F1 distribution that has been generalized by an F2 distribution is,
- [math]\displaystyle{ Y\sim F_1\bigwedge F_N }[/math]
In this instance, it is written as,
- [math]\displaystyle{ Y\sim \operatorname{Pois(\lambda)}\bigwedge \operatorname{Pois(\phi)} }[/math]
Finally, the probability generating function is,
- [math]\displaystyle{ G_Y(z) = \exp(\lambda(e^{\phi (z-1)}-1)) }[/math]
From the generating function of probabilities we can calculate the probability mass function explained below.
Probability mass function
Let X1,X2,...Xj be Poisson independent variables. The probability distribution of the random variable Y = X1 +X2+...Xj is the Neyman's Type A distribution with parameters [math]\displaystyle{ \lambda }[/math] and [math]\displaystyle{ \phi }[/math].
- [math]\displaystyle{ p_x = P(Y=x) = \frac{e^{-\lambda}\phi^x}{x!} \sum_{j=0}^{\infty} \frac{(\lambda e^{-\phi})^j j^x}{j!}~~ }[/math][math]\displaystyle{ ~~x = 1,2,... }[/math]
Alternatively,
- [math]\displaystyle{ p_x = P(Y=x) = \frac{e^{-\lambda + \lambda e^{-\phi}}\phi^x}{x!} \sum_{j=0}^{x}S(x,j)\lambda^j e^{-\phi^j} }[/math]
In order to see how the previous expression develops, we must bear in mind that the probability mass function is calculated from the probability generating function, and use the property of Stirling Numbers. Let's see the development
- [math]\displaystyle{ G(z) = e^{-\lambda + \lambda e^{-\phi}}\sum_{j=0}^{\infty}\frac{\lambda^j e^{-j\phi}(e^{\phi z}-1)^j}{j|} }[/math]
- [math]\displaystyle{ = e^{-\lambda + \lambda e^{-\phi}}\sum_{j=0}^{\infty} (\lambda e^{-\phi})^j \sum_{x=j}^{\infty}\frac{S(x,j)\phi^x z^x}{x!} }[/math]
- [math]\displaystyle{ = \frac{e^{-\lambda + \lambda e^{-\phi}}\phi^x}{x!} \sum_{j=0}^{x}S(x,j)\lambda^j e^{-\phi^j} }[/math]
Another form to estimate the probabilities is with recurring successions,[6]
- [math]\displaystyle{ p_x = P(Y=x) = \frac{\lambda\phi e^{- \phi}}{x} \sum_{r=0}^{x-1} \frac{\phi^r}{r!} p_{x-r-1}~~ }[/math],[math]\displaystyle{ ~~p_0 = \exp(-\lambda + \lambda e ^{- \phi}) }[/math]
Although its length varies directly with n, this recurrence relation is only employed for numerical computation and is particularly useful for computer applications.
where
- x = 0, 1, 2, ... , except for probabilities of recurring successions, where x = 1, 2, 3, ...
- [math]\displaystyle{ j \leq x }[/math]
- [math]\displaystyle{ \lambda }[/math], [math]\displaystyle{ \phi\gt 0 }[/math].
- x! and j! are the factorials of x and j, respectively.
- one of the properties of Stirling numbers of the second kind is as follows:[7]
- [math]\displaystyle{ (e^{\phi z} - 1)^j = j! \sum_{x=j}^\infty \frac{S(x,j){\phi z}^x}{x!} }[/math]
Notation
- [math]\displaystyle{ Y\ \sim \operatorname{NA}(\lambda,\phi)\, }[/math]
Properties
Moment and cumulant generating functions
The moment generating function of a random variable X is defined as the expected value of et, as a function of the real parameter t. For an [math]\displaystyle{ \operatorname{NA(\lambda,\phi)} }[/math], the moment generating function exists and is equal to
- [math]\displaystyle{ M(t) = G_Y(e^t) = \exp(\lambda(e^{\phi (e^t-1)}-1)) }[/math]
The cumulant generating function is the logarithm of the moment generating function and is equal to [1]
- [math]\displaystyle{ K(t) = \log(M(t)) = \lambda(e^{\phi (e^t-1)}-1) }[/math]
In the following table we can see the moments of the order from 1 to 4
| Order | Moment | Cumulant |
|---|---|---|
| 1 | [math]\displaystyle{ \mu_1 = \lambda \phi }[/math] | [math]\displaystyle{ \mu }[/math] |
| 2 | [math]\displaystyle{ \mu_2 = \lambda \phi(1 + \phi) }[/math] | [math]\displaystyle{ \sigma^2 }[/math] |
| 3 | [math]\displaystyle{ \mu_3 = \lambda \phi(1 + 3\phi + \phi^2) }[/math] | [math]\displaystyle{ k_3 }[/math] |
| 4 | [math]\displaystyle{ \mu_4 = \lambda \phi(1 + 7\phi + 6\phi^2 + \phi^3) + 3\lambda^2\phi^2(1 + \phi)^2 }[/math] | [math]\displaystyle{ k_4 + 3\sigma^4 }[/math] |
Skewness
The skewness is the third moment centered around the mean divided by the 3/2 power of the standard deviation, and for the [math]\displaystyle{ \operatorname{NA} }[/math] distribution is,
- [math]\displaystyle{ \gamma_1 = \frac{\mu_3}{\mu_2^{3/2}} = \frac{\lambda \phi(1 + 3\phi + \phi^2)}{(\lambda \phi(1 + \phi))^{3/2}} }[/math]
Kurtosis
The kurtosis is the fourth moment centered around the mean, divided by the square of the variance, and for the [math]\displaystyle{ \operatorname{NA} }[/math] distribution is,
- [math]\displaystyle{ \beta_2= \frac{\mu_4}{\mu_2^2} = \frac{\lambda \phi(1 + 7\phi + 6\phi^2 + \phi^3) + 3\lambda^2\phi^2(1 + \phi)^2}{\lambda^2 \phi^2(1 + \phi)^2} = \frac{1+7\phi+6\phi^2+\phi^3}{\phi\lambda (1+\phi)^2} + 3 }[/math]
The excess kurtosis is just a correction to make the kurtosis of the normal distribution equal to zero, and it is the following,
- [math]\displaystyle{ \gamma_2= \frac{\mu_4}{\mu_2^2}-3 = \frac{1+7\phi+6\phi^2+\phi^3}{\phi\lambda (1+\phi)^2} }[/math]
- Always [math]\displaystyle{ \beta_2 \gt 3 }[/math], or [math]\displaystyle{ \gamma_2 \gt 0 }[/math] the distribution has a high acute peak around the mean and fatter tails.
Characteristic function
In a discrete distribution the characteristic function of any real-valued random variable is defined as the expected value of [math]\displaystyle{ e^{itX} }[/math], where i is the imaginary unit and t ∈ R
- [math]\displaystyle{ \phi(t)= E[e^{itX}] = \sum_{j=0}^\infty e^{ijt}P[X=j] }[/math]
This function is related to the moment generating function via [math]\displaystyle{ \phi_x(t) = M_X(it) }[/math]. Hence for this distribution the characteristic function is,
- [math]\displaystyle{ \phi_x(t) = \exp(\lambda(e^{\phi (e^{it}-1)}-1)) }[/math]
- Note that the symbol [math]\displaystyle{ \phi_x }[/math] is used to represent the characteristic function.
Cumulative distribution function
The cumulative distribution function is,
- [math]\displaystyle{ \begin{align} F(x;\lambda,\phi)& = P(Y \leq x)\\ &= e^{-\lambda} \sum_{i=0}^{x} \frac{\phi^i}{i!} \sum_{j=0}^{\infty} \frac{(\lambda e^{-\phi})^j j^i}{j!}\\ &= e^{-\lambda} \sum_{i=0}^{x} \sum_{j=0}^{\infty} \frac{\phi^i(\lambda e^{-\phi})^j j^i}{i!j!} \end{align} }[/math]
Other properties
- The index of dispersion is a normalized measure of the dispersion of a probability distribution. It is defined as the ratio of the variance [math]\displaystyle{ \sigma^2 }[/math] to the mean [math]\displaystyle{ \mu }[/math],[8]
- [math]\displaystyle{ d = \frac{\sigma^2}{\mu} = 1 + \phi }[/math]
- From a sample of size N, where each random variable Yi comes from a [math]\displaystyle{ \operatorname{NA(\lambda,\phi)} }[/math], where Y1, Y2, .., Yn are independent. This gives the MLE estimator as,[9]
- [math]\displaystyle{ \sum_{i=1}^{N} \frac{x_i}{N} = \bar{x} = \bar{\lambda}\bar{\phi} ~~~ }[/math] where [math]\displaystyle{ \mu }[/math] is the poblational mean of [math]\displaystyle{ \bar{x} }[/math]
- Between the two earlier expressions We are able to parametrize using [math]\displaystyle{ \mu }[/math] and [math]\displaystyle{ d }[/math],
- [math]\displaystyle{ \begin{cases} \mu = \lambda\phi \\ d = 1 + \phi \end{cases} \longrightarrow \quad\! \begin{aligned} \lambda = \frac{\mu}{d-1} \\ \phi = d -1 \end{aligned} }[/math]
Parameter estimation
Method of moments
The mean and the variance of the NA([math]\displaystyle{ \lambda,\phi }[/math]) are [math]\displaystyle{ \mu = \lambda\phi }[/math] and [math]\displaystyle{ \lambda\phi(1 + \phi) }[/math], respectively. So we have these two equations,[10]
- [math]\displaystyle{ \begin{cases} \bar{x} = \lambda\phi \\ s^2 = \lambda\phi(1 + \phi) \end{cases} }[/math]
- [math]\displaystyle{ s^2 }[/math] and [math]\displaystyle{ \bar{x} }[/math] are the mostral variance and mean respectively.
Solving these two equations we get the moment estimators [math]\displaystyle{ \hat{\lambda} }[/math] and [math]\displaystyle{ \hat{\phi} }[/math] of [math]\displaystyle{ \lambda }[/math] and [math]\displaystyle{ \phi }[/math].
- [math]\displaystyle{ \bar{\lambda} = \frac{\bar{x}^2}{s^2 - \bar{x}} }[/math]
- [math]\displaystyle{ \bar{\phi} = \frac{s^2 - \bar{x}}{\bar{x}} }[/math]
Maximum likelihood
Calculating the maximum likelihood estimator of [math]\displaystyle{ \lambda }[/math] and [math]\displaystyle{ \phi }[/math] involves multiplying all the probabilities in the probability mass function to obtain the expression [math]\displaystyle{ \mathcal{L}(\lambda,\phi;x_1,\ldots,x_n) }[/math].
When we apply the parameterization adjustment defined in "Other Properties," we get [math]\displaystyle{ \mathcal{L}(\mu, d;X) }[/math]. We may define the Maximum likelihood estimation based on a single parameter if we estimate the [math]\displaystyle{ \mu }[/math] as the [math]\displaystyle{ \bar{x} }[/math] (sample mean) given a sample X of size N. We can see it below.
- [math]\displaystyle{ \mathcal{L}(d;X)= \prod_{i=1}^n P(x_i;d) }[/math]
- To estimate the probabilities, we will use the p.m.f. of recurring successions, so that the calculation is less complex.
Testing Poisson assumption
When [math]\displaystyle{ \operatorname{NA(\lambda,\phi)} }[/math] is used to simulate a data sample it is important to see if the Poisson distribution fits the data well. For this, the following Hypothesis test is used:
- [math]\displaystyle{ \begin{cases} H_0: d=1 \\ H_1: d\gt 1 \end{cases} }[/math]
Likelihood-ratio test
The likelihood-ratio test statistic for [math]\displaystyle{ \operatorname{NA} }[/math] is,
- [math]\displaystyle{ W = 2(\mathcal{L}(X;\mu,d)-\mathcal{L}(X;\mu,1)) }[/math]
Where likelihood [math]\displaystyle{ \mathcal{L}() }[/math] is the log-likelihood function. W does not have an asymptotic [math]\displaystyle{ \chi_1^2 }[/math] distribution as expected under the null hypothesis since d = 1 is at the parameter domain's edge. In the asymptotic distribution of W, it can be demonstrated that the constant 0 and [math]\displaystyle{ \chi_1^2 }[/math] have a 50:50 mixture. For this mixture, the [math]\displaystyle{ \alpha }[/math] upper-tail percentage points are the same as the [math]\displaystyle{ 2\alpha }[/math] upper-tail percentage points for a [math]\displaystyle{ \chi_1^2 }[/math]
Related distributions
The poisson distribution (on { 0, 1, 2, 3, ... }) is a special case of the Neyman Type A distribution, with
- [math]\displaystyle{ \operatorname{Pois}(\lambda) = \operatorname{NA}(\lambda,\, 0).\, }[/math]
From moments of order 1 and 2 we can write the population mean and variance based on the parameters [math]\displaystyle{ \lambda }[/math] and [math]\displaystyle{ \phi }[/math].
- [math]\displaystyle{ \mu = \lambda\phi }[/math]
- [math]\displaystyle{ \sigma^2 = \lambda\phi (1 + \phi) }[/math]
In the dispersion index d we observe that by substituting [math]\displaystyle{ \mu }[/math] for the parametrized equation of order 1 and [math]\displaystyle{ \sigma }[/math] for the one of order 2, we obtain [math]\displaystyle{ d = 1 + \phi }[/math]. Our variable Y is therefore distributed as a Poisson of parameter [math]\displaystyle{ \lambda }[/math] when d approaches to 1.
Then we have that,
[math]\displaystyle{ \lim_{d\rightarrow 1}\operatorname{NA}(\lambda,\, \phi) \rightarrow \operatorname{Pois}(\lambda) }[/math]
Applications
Usage History
When the Neyman's type A species' reproduction results in clusters, a distribution has been used to characterize the dispersion of plants. This typically occurs when a species develops from offspring of parent plants or from seeds that fall close to the parent plant. However, Archibald (1948][11] observed that there is insufficient data to infer the kind of reproduction from the type of fitted distribution. While Neyman type A produced positive results for plant distributions, Evans (1953][3] showed that Negative binomial distribution produced positive results for insect distributions. Neyman type A distributions have also been studied in the context of ecology, and the results that unless the plant clusters are so compact as to not lie across the edge of the square used to pick sample locations, the distribution is unlikely to be applicable to plant populations. The compactness of the clusters is a hidden assumption in Neyman's original derivation of the distribution, according to Skellam (1958).[12] The results were shown to be significantly impacted by the square size selection.
In the context of bus driver accidents, Cresswell and Froggatt (1963) [4] derived the Neyman type A based on the following hypotheses:
- Each driver is susceptible to "spells," the number of which is Poissonian for any given amount of time, with the same parameter [math]\displaystyle{ \lambda }[/math] for all drivers.
- A driver's performance during a spell is poor, and he is likely to experience a Poissonian number of collisions, with the same parameter [math]\displaystyle{ \phi }[/math] for all drivers.
- Each driver behaves independently.
- No accidents can occur outside of a spell.
These assumptions lead to a Neyman type A distribution via the [math]\displaystyle{ \operatorname{Pois(\lambda)} }[/math] and [math]\displaystyle{ \operatorname{Pois(\phi)} }[/math] model. In contrast to their "short distribution," Cresswell and Froggatt called this one the "long distribution" because of its lengthy tail. According to Irwin(1964), a type A distribution can also be obtained by assuming that various drivers have different levels of proneness, or K. with probability:
- [math]\displaystyle{ \operatorname{P(K = k\phi)} = \frac{e^{-\lambda} \lambda^k}{k!} }[/math]
- taking values [math]\displaystyle{ ~ 0,~ \phi, ~ 2\phi, ~ 3\phi, ~ ... }[/math]
and that a driver with proneness k[math]\displaystyle{ \phi }[/math] has X accidents where:
- [math]\displaystyle{ ~\operatorname{P(X = x|k\phi)} = \frac{e^{-k\phi}(k\phi)^x}{x!} }[/math]
This is the [math]\displaystyle{ ~\operatorname{Pois(k\phi)} \wedge \operatorname{Pois(\lambda)} }[/math] model with mixing over the values taken by K.
Distribution was also suggested in the application for the grouping of minority tents for food from 1965 to 1969. In this regard, it was predicted that only the clustering rates or the average number of entities per grouping needed to be approximated, rather than adjusting distributions on d very big data bases.
- The reference of the usage history.[1]
Calculating Neyman Type A probabilities in R
- The code below simulates 5000 instances of [math]\displaystyle{ \operatorname{NA(\lambda,\phi)} }[/math],
rNeymanA <- function(n,lambda, phi){
r <- numeric()
for (j in 1:n) {
k = rpois(1,lambda)
r[j] <- sum(rpois(k,phi))
}
return(r)
}
- The mass function of recurring probabilities is implemented in R in order to estimate theoretical probabilities; we can see it below,
dNeyman.rec <- function(x, lambda, phi){
p <- numeric()
p[1]<- exp(-lambda + lambda*exp(-phi))
c <- lambda*phi*exp(-phi)
if(x == 0){
return(p[1])
}
else{
for (i in 1:x) {
suma = 0
for (r in 0:(i-1)) {
suma = suma + (phi^r)/(factorial(r))*p[i-r]
}
p[i+1] = (c/(i))*suma # +1 per l'R
}
res <- p[i+1]
return(res)
}
}
We compare results between the relative frequencies obtained by the simulation and the probabilities computed by the p.m.f. . Given two values for the parameters [math]\displaystyle{ \lambda = 2 }[/math] and [math]\displaystyle{ \phi= 1 }[/math]. It is displayed in the following table,
| X | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| Estimated [math]\displaystyle{ f_i }[/math] | .271 | .207 | .187 | .141 | .082 | .052 | .028 | .016 | .008 |
| Teoric [math]\displaystyle{ P(X = x_i) }[/math] | .282 | .207 | .180 | .129 | .084 | .051 | .029 | .016 | .008 |
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Johnson, N.L; Kemp, A.W; Kotz, S (2005). Univariate Discret Distributions. Canada: Wiley-Intersciencie. pp. 407–409. ISBN 0-471-27246-9.
- ↑ Beall, G. (1940). "The fit and significance of contagious distributions when applied to observations on larval insects". Ecology 21 (4): 460–474. doi:10.2307/1930285. Bibcode: 1940Ecol...21..460B.
- ↑ 3.0 3.1 Evans, D.A. (1953). "Experimental evidence concerning contagious distributions in ecology". Biometrika 40 (1–2): 186–211. doi:10.1093/biomet/40.1-2.186.
- ↑ 4.0 4.1 Creswell, W.L.; Froggatt, P. (1963). The Causation of Bus Driver Accidents. London: Oxford University Press.
- ↑ Feller, W. (1943). "On a general class of "contagious" distributions". Annals of Mathematical Statistics 14 (4): 389. doi:10.1214/aoms/1177731359.
- ↑ Johnson, N.L; Kemp, A.W; Kotz, S (2005). Univariate Discret Distributions. Canada: Wiley-Intersciencie. p. 404.
- ↑ Johnson, N.L; Kemp, A.W; Kotz, S (2005). Univariate Discret Distributions. Canada: Wiley-Intersciencie.
- ↑ "Índice de dispersión (Varianza a Razón media)". April 18, 2022. https://statologos.com/indice-de-dispersion/.
- ↑ Shenton, L.R; Bowman, K.O (1977). Maximum Likelihood Estimation in Small Samples. London: Griffin.
- ↑ Shenton, L.R (1949). "On the efficiency of the method of moments and Neyman Type A Distribution". Biometrika 36: 450–454. doi:10.1093/biomet/36.3-4.450. https://www.jstor.org/stable/2332680.
- ↑ Archibald, E.E.A.. Plant populations I: A new application of Neyman's contagious distribution, Annals of Botany. London: Oxford University Press. pp. 221–235.
- ↑ Skellam, J.G. (1958). "On the derivation and applicability of Neyman's type A distribution". Biometrika 45 (1–2): 32–36. doi:10.1093/biomet/45.1-2.32.
 |

