Biology:Evo-devo gene toolkit
The evo-devo gene toolkit is the small subset of genes in an organism's genome whose products control the organism's embryonic development. Toolkit genes are central to the synthesis of molecular genetics, palaeontology, evolution and developmental biology in the science of evolutionary developmental biology (evo-devo). Many of them are ancient and highly conserved among animal phyla.
Toolkit
Toolkit genes are highly conserved among phyla, meaning that they are ancient, dating back to the last common ancestor of bilaterian animals. For example, that ancestor had at least 7 Pax genes for transcription factors.[1]
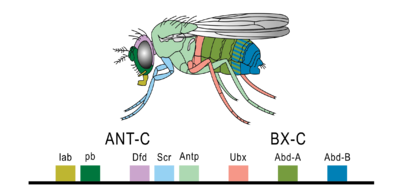
Differences in deployment of toolkit genes affect the body plan and the number, identity, and pattern of body parts. The majority of toolkit genes are components of signaling pathways and encode for the production of transcription factors, cell adhesion proteins, cell surface receptor proteins (and signalling ligands that bind to them), and secreted morphogens; all of these participate in defining the fate of undifferentiated cells, generating spatial and temporal patterns that, in turn, form the body plan of the organism. Among the most important of the toolkit genes are those of the Hox gene cluster, or complex. Hox genes, transcription factors containing the more broadly distributed homeobox protein-binding DNA motif, function in patterning the body axis. Thus, by combinatorially specifying the identity of particular body regions, Hox genes determine where limbs and other body segments will grow in a developing embryo or larva. A paradigmatic toolkit gene is Pax6/eyeless, which controls eye formation in all animals. It has been found to produce eyes in mice and Drosophila, even if mouse Pax6/eyeless was expressed in Drosophila.[2]
This means that a big part of the morphological evolution undergone by organisms is a product of variation in the genetic toolkit, either by the genes changing their expression pattern or acquiring new functions. A good example of the first is the enlargement of the beak in Darwin's large ground-finch (Geospiza magnirostris), in which the gene BMP is responsible for the larger beak of this bird, relative to the other finches.[3]
The loss of legs in snakes and other squamates is another good example of genes changing their expression pattern. In this case the gene Distal-less is very under-expressed, or not expressed at all, in the regions where limbs would form in other tetrapods.[4] In 1994, Sean B. Carroll's team made the "groundbreaking" discovery that this same gene determines the eyespot pattern in butterfly wings, showing that toolbox genes can change their function.[5][6][7]
Toolkit genes, as well as being highly conserved, also tend to evolve the same function convergently or in parallel. Classic examples of this are the already mentioned Distal-less gene, which is responsible for appendage formation in both tetrapods and insects, or, at a finer scale, the generation of wing patterns in the butterflies Heliconius erato and Heliconius melpomene. These butterflies are Müllerian mimics whose coloration pattern arose in different evolutionary events, but is controlled by the same genes.[8] This supports Marc Kirschner and John C. Gerhart's theory of Facilitated Variation, which states that morphological evolutionary novelty is generated by regulatory changes in various members of a large set of conserved mechanisms of development and physiology.[9]
See also
- Endless Forms Most Beautiful
- How the Snake Lost its Legs
References
- ↑ Friedrich, Markus (2015). "Evo-Devo gene toolkit update: at least seven Pax transcription factor subfamilies in the last common ancestor of bilaterian animals Authors". Evolution & Development 17 (5): 255–257. doi:10.1111/ede.12137. PMID 26372059. https://zenodo.org/record/1233833.
- ↑ Xu, P. X.; Woo, I.; Her, H.; Beier, D. R.; Maas, R. L. (1997). "Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode". Development 124 (1): 219–231. doi:10.1242/dev.124.1.219. PMID 9006082.
- ↑ Abzhanov, A.; Protas, M.; Grant, B. R.; Grant, P. R.; Tabin, C. J. (2004). "Bmp4 and Morphological Variation of Beaks in Darwin's Finches". Science 305 (5689): 1462–1465. doi:10.1126/science.1098095. PMID 15353802. Bibcode: 2004Sci...305.1462A.
- ↑ Cohn, M. J.; Tickle, C. (1999). "Developmental basis of limblessness and axial patterning in snakes". Nature 399 (6735): 474–479. doi:10.1038/20944. PMID 10365960. Bibcode: 1999Natur.399..474C.
- ↑ Beldade, P.; Brakefield, P. M.; Long, A.D. (2002). "Contribution of Distal-less to quantitative variation in butterfly eyespots". Nature 415 (6869): 315–318. doi:10.1038/415315a. PMID 11797007.
- ↑ Werner, Thomas (2015). "Leopard Spots and Zebra Stripes on Fruit Fly Wings". Nature Education 8 (2): 3. https://www.nature.com/scitable/topicpage/leopard-spots-and-zebra-stripes-on-fruit-131087142.
- ↑ Carroll, Sean B. (1994). "Pattern formation and eyespot determination in butterfly wings". Science 265 (5168): 109–114. doi:10.1126/science.7912449. PMID 7912449. Bibcode: 1994Sci...265..109C.
- ↑ Baxter, S. W.; Papa, R.; Chamberlain, N.; Humphray, S. J.; Joron, M.; Morrison, C.; ffrench-Constant, R. H.; McMillan, W. O. et al. (2008). "Convergent Evolution in the Genetic Basis of Mullerian Mimicry in Heliconius Butterflies". Genetics 180 (3): 1567–1577. doi:10.1534/genetics.107.082982. PMID 18791259.
- ↑ Gerhart, John; Kirschner, Marc (2007). "The theory of facilitated variation". Proceedings of the National Academy of Sciences 104 (suppl1): 8582–8589. doi:10.1073/pnas.0701035104. PMID 17494755. Bibcode: 2007PNAS..104.8582G.
 |