Biology:C6orf58
 Generic protein structure example |
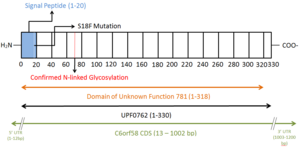
C6orf58 is a humangene located at locus 6q22.33 of chromosome 6 and encodes for UPF0762, a protein which is subsequently secreted after cleavage of a signal peptide.[1] DUF781, which is the singular identifiable domain in UPF0762, is tied to liver development in an orthologous protein in zebrafish.[2] The function of the human UPF0762 is not yet well characterized.[3]
Gene and mRNA
| Genomic DNA Length (base pairs) | Exons | Mature mRNA Length (base pairs) | Splice variants | Signal peptide CDS (base pair) | Mature Peptide CDS (base pair) | 5'-UTR (base pair) | 3'-UTR (base pair) |
|---|---|---|---|---|---|---|---|
| 14644[1] | 6[1] | 1200[1] | 3[3] | 13-72[1] | 73-1002[1] | 1-12[1] | 1003-1200[1] |
Expression
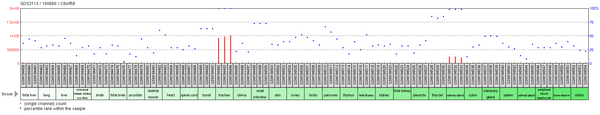
While there are 3 splice variants of C6orf58, only one encodes a good protein.[3] In humans, C6orf58 expressed sequence tags were primarily detected in the larynx and trachea.[4] Transcripts were only detected during the adult stage of development.[4] Experimental microarray data, however, reveals additional regions of C6orf58 expression, namely in the salivary gland, thyroid, and small intestine.[5] Arsenic may also regulate expression as it increases methylation of the C6orf58 promoter.[6]
Gene Neighborhood
Genes within 500 Kilobases of C6orf58 include RSPO3, C6orf174, KIAA0408, RPL17P23, ECHDC1, RPL5P18, YWHAZP4, LOC100420743, LOC100421513, MRPS17P5, and THEMIS.
Homology
A selected set of homologous sequences are listed below, with sequence identity being calculated in comparison to the human reference sequence.
| Species | Common Name | Accession Number | Sequence Length (base pairs) | Sequence Identity |
|---|---|---|---|---|
| Nomascus leucogenys | Northern white-cheeked gibbon | XM_003255689.1 | 1190 | .97 |
| Macaca mulatta | Rhesus monkey | NM_001194318.1 | 1190 | .95 |
| Oryctolagus cuniculus | European rabbit | XM_002714721.1 | 1014 | .79 |
| Loxodonta africana | African bush elephant | XM_003404026.1 | 1020 | .78 |
| Cavia porcellus | Guinea pig | XM_003468475.1 | 1017 | .76 |
| Equus caballus | Horse | XM_001917090.1 | 990 | .77 |
Protein
Properties
| Amino acid length (amino acids) | Signal Peptide Length (amino acids) | Molecular Weight of Precursor Protein | Molecular Weight of Signal Peptide (Predicted) | Molecular Weight of Mature Peptide(predicted) | Molecular Weight(observed) | Isoelectric Point (Predicted) | N-linked glycosylation Site |
|---|---|---|---|---|---|---|---|
| 330[1] | 20[1] | 37.9 kDa[7] | 2.1 kDa[7] | 35.8 kDa[7] | 32 kDa[8] | 5.78[7] | Amino acid 69 |
Mass spectrometry has shown that the observed molecular weight of UPF0762 is 32kDa.[8] It remains unclear why the observed molecular weight is less than predicted, even after accounting for cleavage of the signal peptide. Attachment of a sugar at the site of N-linked glycosylation would also increase the molecular weight.
Homology
UPF0762 shows high homology in primates and orthologous proteins can be traced back as far as trichoplax adhaerens. The list of proteins below is not a comprehensive listing of UPF0762 orthologs. Sequence identity and similarity were determined using BLAST[9] with the reference human sequence as the query.
| Species | Common Name | Accession Number | Sequence Length (amino acids) | Sequence Identity (%) | Sequence Similarity (%) |
|---|---|---|---|---|---|
| Pan troglodytes | Chimpanzee | XP_518733.2 | 330 | 1 | 1 |
| Pongo abelii | Sumatran orangutan | XP_002817388.1 | 330 | .98 | .99 |
| Callithrix jacchus | Marmoset | XP_002746989.1 | 330 | .87 | .93 |
| Canis lupus | Gray wolf | XP_851589.1 | 310 | .7 | .82 |
| Taeniopygia guttata | Zebra finch | XP_002190886.1 | 364 | .43 | .63 |
| Gallus gallus | Red junglefowl | XP_419749.3 | 371 | .42 | .6 |
| Xenopus tropicalis | Western clawed frog | XP_002940437.1 | 178 | 0.29 | 0.51 |
| Trichoplax adhaerens | N/A | XP_002111384 | 381 | .34 | .49 |
Conserved domains
DUF781 is the singular domain of the protein and spans 318 of the protein's 330 amino acids. DUF781 has been linked to liver development in zebrafish.[2]
Post-translational modifications
Observed post-translational modifications include N-linked glycosylation at amino acid 69.[10] A signal peptide, which is predicted to direct the protein to the endoplasmic reticulum for secretion,[11] is cleaved from the first 20 amino acids of the peptide sequence.[1] The missense mutation S18F detected in hepatocellular carcinoma[12] significantly reduces the predicted cleavage score of the signal peptide.[13]
Interactions
Human C6orf58 has been reported to interact with the enzyme ribonucleotide reductase as encoded by the vaccinia virus through a yeast two-hybrid screen.[14]
Pathology
Statistical analysis has shown C6orf58 to be associated with pancreatic cancer survival time.[15] In addition, a missense mutation at amino acid 18 has been observed in liver cancer cells where serine becomes phenylalanine.[12] Analysis of the mutated protein sequence for a signal peptide shows cleavability at the regular amino acid 20 is lost.[13] DUF781's association with liver development and the missense mutation's association with liver cancer is a correlation that remains to be investigated.
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 "Homo sapiens chromosome 6 open reading frame 58 (C6orf58), mRNA". National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/nuccore/NM_001010905.1.
- ↑ 2.0 2.1 "liver-enriched gene 1a and 1b encode novel secretory proteins essential for normal liver development in zebrafish". PLOS ONE 6 (8): e22910. 2011. doi:10.1371/journal.pone.0022910. PMID 21857963. Bibcode: 2011PLoSO...622910C.
- ↑ 3.0 3.1 3.2 Thierry-Mieg, Danielle. "AceView: integrative annotation of cDNA-supported genes in human, mouse, rat, worm and Arabidopsis". NCBI. https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&term=c6orf58&submit=Go.
- ↑ 4.0 4.1 "EST Profile Hs.226268". NCBI. https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.226268.
- ↑ "A comprehensive functional analysis of tissue specificity of human gene expression". BMC Biol. 6: 49. 2008. doi:10.1186/1741-7007-6-49. PMID 19014478.
- ↑ "Epigenetic changes in individuals with arsenicosis". Chem. Res. Toxicol. 24 (2): 165–7. 2011. doi:10.1021/tx1004419. PMID 21291286.
- ↑ 7.0 7.1 7.2 7.3 "Protein identification and analysis tools in the ExPASy server". 2-D Proteome Analysis Protocols. Methods Mol. Biol.. 112. 1999. pp. 531–52. doi:10.1385/1-59259-584-7:531. ISBN 1-59259-584-7. http://web.expasy.org/protparam/. Retrieved 30 April 2012.
- ↑ 8.0 8.1 "Surface integrity governs the proteome of hypomineralized enamel". J. Dent. Res. 89 (10): 1160–5. October 2010. doi:10.1177/0022034510375824. PMID 20651090. https://zenodo.org/record/894336.
- ↑ "Basic local alignment search tool". J. Mol. Biol. 215 (3): 403–10. 1990. doi:10.1016/S0022-2836(05)80360-2. PMID 2231712.
- ↑ "Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry". J. Proteome Res. 5 (6): 1493–503. June 2006. doi:10.1021/pr050492k. PMID 16740002.
- ↑ Caboche, Michel. "Predotar". http://urgi.versailles.inra.fr/predotar/predotar.html.
- ↑ 12.0 12.1 "Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma". Nat. Genet. 43 (9): 828–9. 2011. doi:10.1038/ng.903. PMID 21822264.
- ↑ 13.0 13.1 "SignalP 4.0: discriminating signal peptides from transmembrane regions". Nat. Methods 8 (10): 785–6. 2011. doi:10.1038/nmeth.1701. PMID 21959131.
- ↑ "Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings". J. Proteome Res. 8 (9): 4311–8. 2009. doi:10.1021/pr900491n. PMID 19637933.
- ↑ "A transcriptome analysis by lasso penalized Cox regression for pancreatic cancer survival". J Bioinform Comput Biol 9 Suppl 1: 63–73. 2011. doi:10.1142/s0219720011005744. PMID 22144254.
External links
- Human C6orf58 genome location and C6orf58 gene details page in the UCSC Genome Browser.