Literature-based discovery
Literature-based discovery (LBD), also called literature-related discovery (LRD) is a form of knowledge extraction and automated hypothesis generation that uses papers and other academic publications (the "literature") to find new relationships between existing knowledge (the "discovery"). Literature-based discovery aims to discover new knowledge by connecting information which have been explicitly stated in literature to deduce connections which have not been explicitly stated.[1]
LBD can help researchers to quickly discover and explore hypotheses as well as gain information on relevant advances inside and outside of their niches and increase interdisciplinary information sharing.[1]
The most basic and widespread type of LBD is called the ABC paradigm because it centers around three concepts called A, B and C.[2][3][4] It states that if there is a connection between A and B and one between B and C, then there is one between A and C which, if not explicitly stated, is yet to be explored.[1]
History
The LBD technique was pioneered by Don R. Swanson in the 1980s.[5] He hypothesized that the combination of two separately published results indicating an A-B relationship and a B-C relationship are evidence of an A-C relationship which is unknown or unexplored. He used this to propose fish oil as a treatment for Raynaud syndrome due to their shared relationship with blood viscosity.[6] This hypothesis was later shown to have merit in a prospective study [7] and he continually proposed other discoveries using similar methods.[8][9][10][1]
Swanson linking
Swanson linking is a term proposed in 2003[11] that refers to connecting two pieces of knowledge previously thought to be unrelated.[12] For example, it may be known that illness A is caused by chemical B, and that drug C is known to reduce the amount of chemical B in the body. However, because the respective articles were published separately from one another (called "disjoint data"), the relationship between illness A and drug C may be unknown. Swanson linking aims to find these relationships and report them.
Although the ABC paradigm is widely used, critics of the system have argued that much of science is not captured on simple assertions and it is rather built from analogies and images at a higher level of abstraction.[13]
Systems
LBD comes generally in two flavours: open and closed discovery. In open discovery, only A is given. The approach finds Bs and uses them to return possibly interesting Cs to the user, thus generating hypotheses from A. With closed discovery, the A and C are given to the approach which seeks to find the Bs which can link the two, thus testing a hypothesis about A and C.[1]
A number of systems to perform literature-based discovery have been developed over the years, extending the original idea of Don Swanson, and the evaluation of the quality of such systems is an active area of research.[14] Some systems include web versions for increased user-friendliness.[15] A common approach to many systems is the use of MeSH terms to represent scientific articles. This is used by the systems Manjal, BITOLA and LitLinker.[16]
One well-known system within the field is called Arrowsmith and is tailored to find connections between two disjoint sets of articles, an approach labeled "two-node" search.[17][18]
Another well-known system, LION LBD,[19] uses PubTator [20] for annotating PubMed scientific articles with concepts such as chemicals, genes/proteins, mutations, diseases and species; as well as sentence-level annotation of cancer hallmarks that describe fundamental cancer processes and behaviour.[21] It uses co-occurrence metrics to rank relations between concepts and performs both open and closed discovery.[1]
While LBD systems are based on traditional statistical methods,[16] other systems leverage sophisticated machine learning methods, like neural networks.[1] Some LBD systems represent the connection between concepts as a knowledge graph, and thus employ techniques of graph theory.[22] The graph-based representation is also the foundation for LBD systems that employ graph databases like Neo4J, enabling discovery via graph query languages such as Cypher.[23]
Graph-based LBD systems represent the relations between concepts using a different relation types, such as those in the UMLS Semantic Network.[24] Some approaches go further and try to apply contextualized relations,[25] an approach also used by the Gene Ontology for their Causal Activity Modeling (GO-CAM).[26]
Use of databases
Besides extracting information from the body of scientific articles, LBD systems often employ structured knowledge from biocurated biological resources, like the Online Mendelian Inheritance in Men (OMIM).[27]
List of systems
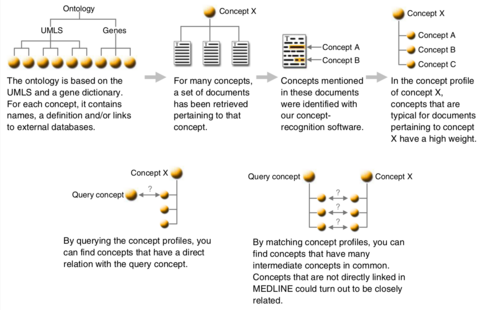
These are the published LBD systems, ordered by date of publication:[29]
- 1986 - Arrowsmith [6]
- 2000 - BITOLA V1 [30]
- 2001 - DAD [31]
- 2003 - LitLinker [32]
- 2004 - ACS [33]
- 2004 - Manjal [34]
- 2004 - IRIDESCENT [35]
- 2005 - BITOLA V2 [36]
- 2006 - LitLinker V2 [37]
- 2007 - Arrowsmith V2 [38]
- 2008 - Anni 2.0 [28]
- 2008 - CoPub Discovery [39]
- 2009 - RajoLink [40]
- 2010 - Sem-BT [41]
- 2015 - Obvio [42]
- 2016 - Spark [43]
- 2017 - Mine the gap [44]
- 2019 - LION LBD [19]
Semantic typing
A common task in literature-based discovery is assigning words/concepts to different semantic types. A concept might be classified under one type or multiple types. For example in the Unified Medical Language System (UMLS) the term migraine is classified under the type disease and syndrome, while the term magnesium is under two types: biologically active substance and element, ion, or isotope.[16] The typing of concepts hones the discovery of connections between particular classes of concepts, i.e. diseases-genes or diseases-drugs. [16]
System evaluation
The evaluation of literature-based discoveries is challenging, and includes both experimental and in silico methods.[45] Methods try to quantify the amount of knowledge generated by systems, that should be provided in an amount and richness that is useful for scientists.[46]
Evaluation is difficult in LBD for several reasons: disagreement about the role of LBD systems in research and thus what makes a successful one; difficulty in determining how useful, interesting or actionable a discovery is; and difficulty in objectively defining a ‘discovery’, which hinders the creation of a standard evaluation set which quantifies when a discovery has been replicated or found.[1]
A popular method used in LBD is to replicate previous discoveries. [4][47][48] These are usually LBD-based discoveries as they are relatively easy to quantify compared to other discoveries. There are only a handful of such discoveries and approaches e tuned to perform well on these discoveries might not generalise. In this type of evaluation, the literature before the discovery to be replicated is used to generate a ranked list of discovery candidates as target or linking terms. Success is measured by reporting the rank of the term(s) of interest; the higher the rank, the better the approach.
Literature- or time-slicing involves splitting the existing literature at a point in time. The LBD system is then exposed to the literature before the split and is evaluated by how many of the discoveries in the later period it can discover. LBD systems have used term co-occurrences,[49] relationships from external biomedical resources (e.g SemMedDB)[50] and semantic relationships[51] to generate the gold standards. A high precision approach is to get expert opinion to generate the gold standard,[52] but this is time-consuming, expensive and tends to produce low recall rates.[1]
The advantage of time-slicing in comparison to the replication of previous discoveries is the evaluation on a large number of test instances. This raises the need for evaluation metrics which can quantify performance on large, ranked lists.[1] LBD works have used metrics popular in Information Retrieval [53] which include Precision, Recall, Area Under the Curve (AUC), Precision at k, Mean Average Precision (MAP) and others.[1]
The approach of Proposing new discoveries or treatments goes beyond replicating past discoveries or predicting time-sliced instances of a particular relationship and shows that a system is capable of being used in realistic situations.[54][47][55][56] This is usually accompanied by peer-reviewed publication in the domain or vetting by a domain expert.[1]
Text mining
The automation of literature-based discovery relies heavily on text mining.[58]
The language in scientific articles often include ambiguities, and an important step for coeherent parsing of the literature is the extraction of the sense of each term in the context they are used, a task called Word-sense disambiguation (WSD).[59] For example, terms for genes like CT (PCYT1A) called and MR (NR3C2) can be confused with the acronyms for Computational Tomography and Magnetic Resonance, requiring sofisticated disambiguation systems.[60] Terms are often reconciled to ontologies or other sources of unique identifiers, such as the Unified Medical Language System (UMLS).[61] This process of mapping multiple different utterances to a single name or identifier is called normalization.[57]
Usage
Life sciences
LBD has already been used in different ways to identify new connections between biomedical entities and new candidate genes and treatments for illnesses.[62][1]
Drug discovery
LBD has seen use in drug development and repurposing [54][63] as well as predicting adverse drug reactions.[64][65][1]
The method of literature-based discovery has been used to search for treatments for a number of human diseases, including:
- diabetic retinopathy[66]
- dilated cardiomyopathy[61]
- Parkinson's disease[67]
- prostate cancer[63]
- gastric cancer[68]
- multiple sclerosis[69]
Gene and protein function discovery
The approach has also been used to propose relations of genes with particular diseases,[70] like breast cancer.[71]
In the context of systems vaccinology, it was used to identify proteins related to interferon gamma and that play a role in the response to vaccines.[57]
It has also been used to propose mechanisms for currently used drugs.[72]
Biomarker discovery
LBD has been explored as a tool to identify biomarkers for diagnostic and prognostic for diseases, e.g. for the risk of type 2 diabetes.[73]
Other uses
Besides providing scientific hypotheses about the world, LBD has also been used to improve data analysis, via the automatic identification of possible confounding factors using the medical literature.[74]
It has also been used to understand better disease etiology and the relation of different diseases, for example looking for the genes connecting myocardial infarction and depression,[75] and connections between psychiatric and somatic diseases.[76]
Beyond life sciences
LBD has mostly been deployed in the biomedical domain, but it has also been used outside of it as it has been applied to research into developing water purification systems, accelerating development of developing countries and identifying promising research collaborations.[77][78][79]
See also
- Arrowsmith System
- Implicature
- Latent semantic indexing
- Metaphor
- Text mining
- Biocuration
- BioCreative
Additional reading
- Wilson, Patrick (1977). Public Knowledge, Private Ignorance: Toward a Library and Information Policy. Greenwood Publishing Group. p. 156. ISBN:0-8371-9485-7.
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Crichton, Gamal; Baker, Simon; Guo, Yufan; Korhonen, Anna (2020-05-15). "Neural networks for open and closed Literature-based Discovery". PLOS ONE 15 (5): e0232891. doi:10.1371/JOURNAL.PONE.0232891. PMID 32413059. PMC 7228051. Bibcode: 2020PLoSO..1532891C. https://www.wikidata.org/wiki/Q94937526.
 This article incorporates text available under the CC BY 4.0 license.
This article incorporates text available under the CC BY 4.0 license.
- ↑ Smalheiser, Neil R; Swanson, Don R (November 1998). "Using Arrowsmith: a computer-assisted approach to formulating and assessing scientific hypotheses". Computer Methods and Programs in Biomedicine 57 (3): 149–153. doi:10.1016/s0169-2607(98)00033-9. ISSN 0169-2607. PMID 9822851. http://dx.doi.org/10.1016/s0169-2607(98)00033-9.
- ↑ Gordon, Michael D.; Lindsay, Robert K. (February 1996). <116::aid-asi3>3.0.co;2-1 "Toward discovery support systems: A replication, re-examination, and extension of Swanson's work on literature-based discovery of a connection between Raynaud's and fish oil". Journal of the American Society for Information Science 47 (2): 116–128. doi:10.1002/(sici)1097-4571(199602)47:2<116::aid-asi3>3.0.co;2-1. ISSN 0002-8231. http://dx.doi.org/10.1002/(sici)1097-4571(199602)47:2<116::aid-asi3>3.0.co;2-1.
- ↑ 4.0 4.1 Cohen, Trevor; Schvaneveldt, Roger; Widdows, Dominic (April 2010). "Reflective Random Indexing and indirect inference: A scalable method for discovery of implicit connections". Journal of Biomedical Informatics 43 (2): 240–256. doi:10.1016/j.jbi.2009.09.003. ISSN 1532-0464. PMID 19761870.
- ↑ Smalheiser, Neil R. (2017-12-01). "Rediscovering Don Swanson:The Past, Present and Future of Literature-based Discovery" (in en). Journal of Data and Information Science 2 (4): 43–64. doi:10.1515/jdis-2017-0019. PMID 29355246.
- ↑ 6.0 6.1 Swanson, Don R. (1986). "Fish Oil, Raynaud's Syndrome, and Undiscovered Public Knowledge". Perspectives in Biology and Medicine 30 (1): 7–18. doi:10.1353/pbm.1986.0087. ISSN 1529-8795. PMID 3797213. http://dx.doi.org/10.1353/pbm.1986.0087.
- ↑ Ricco, Jean Baptiste (May 1990). "Fish-oil dietary supplementation in patients with Raynaud's phenomenon: a double blind, controlled, prospective study". Journal of Vascular Surgery 11 (5): 733–734. doi:10.1016/0741-5214(90)90229-4. ISSN 0741-5214.
- ↑ Swanson, Don R. (1988). "Migraine and Magnesium: Eleven Neglected Connections". Perspectives in Biology and Medicine 31 (4): 526–557. doi:10.1353/pbm.1988.0009. ISSN 1529-8795. PMID 3075738. http://dx.doi.org/10.1353/pbm.1988.0009.
- ↑ Swanson, Don R. (1990). "Somatomedin C and Arginine: Implicit Connections between Mutually Isolated Literatures". Perspectives in Biology and Medicine 33 (2): 157–186. doi:10.1353/pbm.1990.0031. ISSN 1529-8795. PMID 2406696. http://dx.doi.org/10.1353/pbm.1990.0031.
- ↑ Smalheiser, Neil R.; Swanson, Don R. (September 1996). "Linking estrogen to Alzheimer's disease". Neurology 47 (3): 809–810. doi:10.1212/wnl.47.3.809. ISSN 0028-3878. PMID 8797484. http://dx.doi.org/10.1212/wnl.47.3.809.
- ↑ Stegmann J, Grohmann G. Hypothesis generation guided by co-word clustering. Scientometrics. 2003;56:111–135. As quoted by Bekhuis
- ↑ Bekhuis, Tanja (2006). "Conceptual biology, hypothesis discovery, and text mining: Swanson's legacy". Biomedical Digital Libraries 3: 2. doi:10.1186/1742-5581-3-2. PMID 16584552.
- ↑ Smalheiser, Neil R. (2011-07-26). "Literature-based discovery: Beyond the ABCs". Journal of the Association for Information Science and Technology 63 (2): 218–224. doi:10.1002/ASI.21599. https://www.wikidata.org/wiki/Q57372419.
- ↑ Yetisgen-Yildiz, Meliha; Pratt, Wanda (2008-12-16). "A new evaluation methodology for literature-based discovery systems.". Journal of Biomedical Informatics 42 (4): 633–643. doi:10.1016/J.JBI.2008.12.001. PMID 19124086. https://www.wikidata.org/wiki/Q51857372.
- ↑ Hur, Junguk; Schuyler, Adam D.; States, David J.; Feldman, Eva L. (2009-02-02). "SciMiner: web-based literature mining tool for target identification and functional enrichment analysis". Bioinformatics 25 (6): 838–840. doi:10.1093/bioinformatics/btp049. ISSN 1460-2059. PMID 19188191. PMC 2654801. http://dx.doi.org/10.1093/bioinformatics/btp049.
- ↑ 16.0 16.1 16.2 16.3 Yetisgen-Yildiz, Meliha; Pratt, Wanda (2006-01-04). "Using statistical and knowledge-based approaches for literature-based discovery.". Journal of Biomedical Informatics 39 (6): 600–611. doi:10.1016/J.JBI.2005.11.010. PMID 16442852. https://www.wikidata.org/wiki/Q51953854.
- ↑ Smalheiser, Neil R.; Torvik, Vetle I. (2008), Bruza, Peter; Weeber, Marc, eds., "The Place of Literature-Based Discovery in Contemporary Scientific Practice" (in en), Literature-based Discovery, Information Science and Knowledge Management (Berlin, Heidelberg: Springer): pp. 13–22, doi:10.1007/978-3-540-68690-3_2, ISBN 978-3-540-68690-3, Bibcode: 2008lbd..book...13S, https://doi.org/10.1007/978-3-540-68690-3_2, retrieved 2022-03-04
- ↑ "ARROWSMITH: Start". http://arrowsmith.psych.uic.edu/cgi-bin/arrowsmith_uic/start.cgi.
- ↑ 19.0 19.1 Pyysalo, Sampo; Baker, Simon; Ali, Imran; Haselwimmer, Stefan; Shah, Tejas; Young, Andrew; Guo, Yufan; Högberg, Johan et al. (2018-10-09). "LION LBD: a literature-based discovery system for cancer biology". Bioinformatics 35 (9): 1553–1561. doi:10.1093/bioinformatics/bty845. ISSN 1367-4803. PMID 30304355. PMC 6499247. http://dx.doi.org/10.1093/bioinformatics/bty845.
- ↑ Wei, Chih-Hsuan; Kao, Hung-Yu; Lu, Zhiyong (2013-05-22). "PubTator: a web-based text mining tool for assisting biocuration". Nucleic Acids Research 41 (W1): W518–W522. doi:10.1093/nar/gkt441. ISSN 1362-4962. PMID 23703206. PMC 3692066. http://dx.doi.org/10.1093/nar/gkt441.
- ↑ Baker, Simon; Ali, Imran; Silins, Ilona; Pyysalo, Sampo; Guo, Yufan; Högberg, Johan; Stenius, Ulla; Korhonen, Anna (2017-07-14). "Cancer Hallmarks Analytics Tool (CHAT): a text mining approach to organize and evaluate scientific literature on cancer". Bioinformatics 33 (24): 3973–3981. doi:10.1093/bioinformatics/btx454. ISSN 1367-4803. PMID 29036271. PMC 5860084. http://dx.doi.org/10.1093/bioinformatics/btx454.
- ↑ Cameron, Delroy; Kavuluru, Ramakanth; Rindflesch, Thomas C.; Sheth, Amit P.; Thirunarayan, Krishnaprasad; Bodenreider, Olivier (2015-02-07). "Context-driven automatic subgraph creation for literature-based discovery". Journal of Biomedical Informatics 54: 141–157. doi:10.1016/J.JBI.2015.01.014. PMID 25661592. PMC 4888806. https://www.wikidata.org/wiki/Q35557700.
- ↑ Hristovski, Dimitar; Kastrin, Andrej; Dinevski, Dejan; Rindflesch, Thomas C. (2015-01-01). "Constructing a Graph Database for Semantic Literature-Based Discovery". Studies in Health Technology and Informatics 216: 1094. PMID 26262393. https://www.wikidata.org/wiki/Q35742372.
- ↑ Preiss, Judita; Stevenson, Mark; Gaizauskas, Robert (2015-05-13). "Exploring relation types for literature-based discovery.". Journal of the American Medical Informatics Association 22 (5): 987–992. doi:10.1093/JAMIA/OCV002. PMID 25971437. PMC 4986660. https://www.wikidata.org/wiki/Q37179375.
- ↑ Kim, Yong Hwan; Song, Min (2019-04-24). "A context-based ABC model for literature-based discovery" (in English). PLOS ONE 14 (4): e0215313. doi:10.1371/JOURNAL.PONE.0215313. PMID 31017923. PMC 6481912. Bibcode: 2019PLoSO..1415313K. https://www.wikidata.org/wiki/Q64095142.
- ↑ Thomas, Paul D.; Hill, David P.; Mi, Huaiyu; Osumi-Sutherland, David; Auken, Kimberly Van; Carbon, Seth J.; Balhoff, James P.; Albou, Laurent-Philippe et al. (2019-10-01). "Gene Ontology Causal Activity Modeling (GO-CAM) moves beyond GO annotations to structured descriptions of biological functions and systems". Nature Genetics 51 (10): 1429–1433. doi:10.1038/S41588-019-0500-1. PMID 31548717. PMC 7012280. https://www.wikidata.org/wiki/Q90243389.
- ↑ Hristovski, Dimitar; Peterlin, Borut; Mitchell, Joyce A.; Humphrey, Susanne M. (2003-01-01). "Improving literature based discovery support by genetic knowledge integration". Studies in Health Technology and Informatics 95: 68–73. PMID 14663965. https://www.wikidata.org/wiki/Q52006778.
- ↑ 28.0 28.1 Jelier, Rob; Schuemie, Martijn J.; Schuemie, Martijn J.; Veldhoven, Antoine; Dorssers, Lambert C. J.; Jenster, Guido; Kors, Jan A.; Kors, Jan A. (2008-06-12). "Anni 2.0: a multipurpose text-mining tool for the life sciences." (in English). Genome Biology 9 (6): R96. doi:10.1186/GB-2008-9-6-R96. PMID 18549479. PMC 2481428. https://www.wikidata.org/wiki/Q36787149.
- ↑ Gopalakrishnan, Vishrawas; Jha, Kishlay; Jin, Wei; Zhang, Aidong (2019-05-01). "A survey on literature based discovery approaches in biomedical domain" (in en). Journal of Biomedical Informatics 93: 103141. doi:10.1016/j.jbi.2019.103141. ISSN 1532-0464. PMID 30857950.
- ↑ Hristovski, Dimitar; Džeroski, Sašo; Peterlin, Borut; Rožić, Anamajirja (2000), "Supporting Discovery in Medicine by Association Rule Mining of Bibliographic Databases", Principles of Data Mining and Knowledge Discovery (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 446–451, doi:10.1007/3-540-45372-5_49, ISBN 978-3-540-41066-9
- ↑ Weeber, Marc; Klein, Henny; de Jong-van den Berg, Lolkje T.W.; Vos, Rein (2001). "Using concepts in literature-based discovery: Simulating Swanson's Raynaud-fish oil and migraine-magnesium discoveries". Journal of the American Society for Information Science and Technology 52 (7): 548–557. doi:10.1002/asi.1104. ISSN 1532-2882. http://dx.doi.org/10.1002/asi.1104.
- ↑ Pratt, Wanda; Yetisgen-Yildiz, Meliha (2003). "LitLinker". Proceedings of the 2nd international conference on Knowledge capture. New York, New York, USA: ACM Press. p. 105. doi:10.1145/945645.945662. ISBN 1581135831. http://dx.doi.org/10.1145/945645.945662.
- ↑ van der Eijk, C. Christiaan; van Mulligen, Erik M.; Kors, Jan A.; Mons, Barend; van den Berg, Jan (2004). "Constructing an associative concept space for literature-based discovery". Journal of the American Society for Information Science and Technology 55 (5): 436–444. doi:10.1002/asi.10392. ISSN 1532-2882. http://dx.doi.org/10.1002/asi.10392.
- ↑ Srinivasan, P.; Libbus, B. (2004-07-19). "Mining MEDLINE for implicit links between dietary substances and diseases". Bioinformatics 20 (Suppl 1): i290–i296. doi:10.1093/bioinformatics/bth914. ISSN 1367-4803. PMID 15262811. http://dx.doi.org/10.1093/bioinformatics/bth914.
- ↑ Wren, Jonathan D (2004). "Extending the mutual information measure to rank inferred literature relationships". BMC Bioinformatics 5 (1): 145. doi:10.1186/1471-2105-5-145. PMID 15471547.
- ↑ Hristovski, Dimitar; Peterlin, Borut; Mitchell, Joyce A.; Humphrey, Susanne M. (March 2005). "Using literature-based discovery to identify disease candidate genes". International Journal of Medical Informatics 74 (2–4): 289–298. doi:10.1016/j.ijmedinf.2004.04.024. ISSN 1386-5056. PMID 15694635. http://dx.doi.org/10.1016/j.ijmedinf.2004.04.024.
- ↑ Yetisgen-Yildiz, Meliha; Pratt, Wanda (December 2006). "Using statistical and knowledge-based approaches for literature-based discovery". Journal of Biomedical Informatics 39 (6): 600–611. doi:10.1016/j.jbi.2005.11.010. ISSN 1532-0464. PMID 16442852.
- ↑ Torvik, Vetle I.; Smalheiser, Neil R. (2007-04-26). "A quantitative model for linking two disparate sets of articles in MEDLINE". Bioinformatics 23 (13): 1658–1665. doi:10.1093/bioinformatics/btm161. ISSN 1460-2059. PMID 17463015.
- ↑ Frijters, R.; Heupers, B.; van Beek, P.; Bouwhuis, M.; van Schaik, R.; de Vlieg, J.; Polman, J.; Alkema, W. (2008-05-19). "CoPub: a literature-based keyword enrichment tool for microarray data analysis". Nucleic Acids Research 36 (Web Server): W406–W410. doi:10.1093/nar/gkn215. ISSN 0305-1048. PMID 18442992. PMC 2447728. http://dx.doi.org/10.1093/nar/gkn215.
- ↑ Petriĕ, Ingrid; Urbanĕiĕ, Tanja; Cestnik, Bojan; Macedoni-Lukšiĕ, Marta (April 2009). "Literature mining method RaJoLink for uncovering relations between biomedical concepts". Journal of Biomedical Informatics 42 (2): 219–227. doi:10.1016/j.jbi.2008.08.004. ISSN 1532-0464. PMID 18771753.
- ↑ Hristovski, Dimitar; Kastrin, Andrej; Peterlin, Borut; Rindflesch, Thomas C. (2010), "Combining Semantic Relations and DNA Microarray Data for Novel Hypotheses Generation", Linking Literature, Information, and Knowledge for Biology (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 53–61, doi:10.1007/978-3-642-13131-8_7, ISBN 978-3-642-13130-1, http://dx.doi.org/10.1007/978-3-642-13131-8_7, retrieved 2022-03-17
- ↑ Cameron, Delroy; Kavuluru, Ramakanth; Rindflesch, Thomas C.; Sheth, Amit P.; Thirunarayan, Krishnaprasad; Bodenreider, Olivier (April 2015). "Context-driven automatic subgraph creation for literature-based discovery". Journal of Biomedical Informatics 54: 141–157. doi:10.1016/j.jbi.2015.01.014. ISSN 1532-0464. PMID 25661592. PMC 4888806. http://dx.doi.org/10.1016/j.jbi.2015.01.014.
- ↑ Workman, T. Elizabeth; Fiszman, Marcelo; Cairelli, Michael J.; Nahl, Diane; Rindflesch, Thomas C. (2016-04-01). "Spark, an application based on Serendipitous Knowledge Discovery" (in en). Journal of Biomedical Informatics 60: 23–37. doi:10.1016/j.jbi.2015.12.014. ISSN 1532-0464. PMID 26732995.
- ↑ Peng, Yufang; Bonifield, Gary; Smalheiser, Neil R. (2017-05-22). "Gaps within the Biomedical Literature: Initial Characterization and Assessment of Strategies for Discovery". Frontiers in Research Metrics and Analytics 2. doi:10.3389/frma.2017.00003. ISSN 2504-0537. PMID 29271976.
- ↑ Henry, M. S. Sam; McInnes, Bridget T. (2017-08-21). "Literature Based Discovery: models, methods, and trends.". Journal of Biomedical Informatics 74: 20–32. doi:10.1016/J.JBI.2017.08.011. PMID 28838802. https://www.wikidata.org/wiki/Q38371706.
- ↑ Preiss, Judita; Stevenson, Mark (2017-05-31). "Quantifying and filtering knowledge generated by literature based discovery". BMC Bioinformatics 18 (Suppl 7): 249. doi:10.1186/S12859-017-1641-9. PMID 28617217. PMC 5471938. https://www.wikidata.org/wiki/Q33802199.
- ↑ 47.0 47.1 Swanson, Don R.; Smalheiser, Neil R. (April 1997). "An interactive system for finding complementary literatures: a stimulus to scientific discovery". Artificial Intelligence 91 (2): 183–203. doi:10.1016/s0004-3702(97)00008-8. ISSN 0004-3702.
- ↑ R., Weeber, M. Klein, H. Aronson, A. R. Mork, J. G. de Jong-van den Berg, L. T. Vos (2000). "Text-based discovery in biomedicine: the architecture of the DAD-system.". Proceedings. AMIA Symposium (American Medical Informatics Association): 903–907. OCLC 678976989. PMID 11080015. PMC 2243779. http://worldcat.org/oclc/678976989.
- ↑ Hristovski, Dimitar; Džeroski, Sašo; Peterlin, Borut; Rožić, Anamajirja (2000), "Supporting Discovery in Medicine by Association Rule Mining of Bibliographic Databases", Principles of Data Mining and Knowledge Discovery (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 446–451, doi:10.1007/3-540-45372-5_49, ISBN 978-3-540-41066-9
- ↑ Eronen, Lauri; Hintsanen, Petteri; Toivonen, Hannu (2012), "Biomine: A Network-Structured Resource of Biological Entities for Link Prediction", Bisociative Knowledge Discovery (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 364–378, doi:10.1007/978-3-642-31830-6_26, ISBN 978-3-642-31829-0
- ↑ Preiss, Judita; Stevenson, Mark; Gaizauskas, Robert (2015-05-12). "Exploring relation types for literature-based discovery". Journal of the American Medical Informatics Association 22 (5): 987–992. doi:10.1093/jamia/ocv002. ISSN 1527-974X. PMID 25971437. PMC 4986660. http://dx.doi.org/10.1093/jamia/ocv002.
- ↑ Yetisgen-Yildiz, Meliha; Pratt, Wanda (August 2009). "A new evaluation methodology for literature-based discovery systems". Journal of Biomedical Informatics 42 (4): 633–643. doi:10.1016/j.jbi.2008.12.001. ISSN 1532-0464. PMID 19124086.
- ↑ Yetisgen-Yildiz, M.; Pratt, W. (2008), "Evaluation of Literature-Based Discovery Systems", Literature-based Discovery (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 101–113, doi:10.1007/978-3-540-68690-3_7, ISBN 978-3-540-68685-9, Bibcode: 2008lbd..book..101Y, http://dx.doi.org/10.1007/978-3-540-68690-3_7, retrieved 2022-03-15
- ↑ 54.0 54.1 Hristovski, Dimitar; Kastrin, Andrej; Peterlin, Borut; Rindflesch, Thomas C. (2010), "Combining Semantic Relations and DNA Microarray Data for Novel Hypotheses Generation", Linking Literature, Information, and Knowledge for Biology (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 53–61, doi:10.1007/978-3-642-13131-8_7, ISBN 978-3-642-13130-1, http://dx.doi.org/10.1007/978-3-642-13131-8_7, retrieved 2022-03-15
- ↑ Stegmann, Johannes; Grohmann, Guenter (2003). "Hypothesis generation guided by co-word clustering". Scientometrics 56 (1): 111–135. doi:10.1023/A:1021954808804. http://link.springer.com/10.1023/A:1021954808804.
- ↑ Wren, J. D.; Bekeredjian, R.; Stewart, J. A.; Shohet, R. V.; Garner, H. R. (2004-01-22). "Knowledge discovery by automated identification and ranking of implicit relationships". Bioinformatics 20 (3): 389–398. doi:10.1093/bioinformatics/btg421. ISSN 1367-4803. PMID 14960466.
- ↑ 57.0 57.1 57.2 Ozgür, Arzucan; Xiang, Zuoshuang; Radev, Dragomir R.; He, Yongqun (2010-06-03). "Literature-based discovery of IFN-gamma and vaccine-mediated gene interaction networks". Journal of Biomedicine and Biotechnology 2010: 426479. doi:10.1155/2010/426479. PMID 20625487. PMC 2896678. https://www.wikidata.org/wiki/Q33960059.
- ↑ Korhonen, Anna; Guo, Yufan; Baker, Simon; Yetisgen-Yildiz, Meliha; Stenius, Ulla; Narita, Masashi; Liò, Pietro (2015-01-01). "Improving Literature-Based Discovery with Advanced Text Mining" (in English). Computational Intelligence Methods for Bioinformatics and Biostatistics. Lecture Notes in Computer Science. 8623. pp. 89–98. doi:10.1007/978-3-319-24462-4_8. ISBN 978-3-319-24461-7. https://www.wikidata.org/wiki/Q57580185.
- ↑ Preiss, Judita; Stevenson, Mark (July 2016). "The effect of word sense disambiguation accuracy on literature based discovery". BMC Medical Informatics and Decision Making 16 (S1): 57. doi:10.1186/s12911-016-0296-1. ISSN 1472-6947. PMID 27455071.
- ↑ Kastrin, Andrej; Hristovski, Dimitar (2008-11-06). "A fast document classification algorithm for gene symbol disambiguation in the BITOLA literature-based discovery support system". AMIA Annual Symposium Proceedings 2008: 358–362. PMID 18998999. PMC 2655979. https://www.wikidata.org/wiki/Q38511892.
- ↑ 61.0 61.1 Gabetta, Matteo; Larizza, Cristiana; Bellazzi, Riccardo (2013-01-01). "A Unified Medical Language System (UMLS) based system for Literature-Based Discovery in medicine.". Studies in Health Technology and Informatics 192: 412–416. PMID 23920587. https://www.wikidata.org/wiki/Q38447089.
- ↑ Hristovski, Dimitar; Rindflesch, Thomas; Peterlin, Borut (2013-01-01). "Using Literature-based Discovery to Identify Novel Therapeutic Approaches". Cardiovascular & Hematological Agents in Medicinal Chemistry 11 (1): 14–24. doi:10.2174/1871525711311010005. ISSN 1871-5257. PMID 22845900. http://dx.doi.org/10.2174/1871525711311010005.
- ↑ 63.0 63.1 Zhang, Rui; Cairelli, Michael J.; Fiszman, Marcelo; Kilicoglu, Halil; Rindflesch, Thomas C.; Pakhomov, Serguei V.; Melton, Genevieve B. (January 2014). "Exploiting Literature-derived Knowledge and Semantics to Identify Potential Prostate Cancer Drugs". Cancer Informatics 13s1 (Suppl 1): 103–111. doi:10.4137/cin.s13889. ISSN 1176-9351. PMID 25392688. PMC 4216049. http://dx.doi.org/10.4137/cin.s13889.
- ↑ Benzschawel, Eric (2016). "Identifying Potential Adverse Drug Events in Tweets Using Bootstrapped Lexicons". Proceedings of the ACL 2016 Student Research Workshop (Stroudsburg, PA, USA: Association for Computational Linguistics): 15–21. doi:10.18653/v1/p16-3003.
- ↑ Shang, Ning; Xu, Hua; Rindflesch, Thomas C.; Cohen, Trevor (December 2014). "Identifying plausible adverse drug reactions using knowledge extracted from the literature". Journal of Biomedical Informatics 52: 293–310. doi:10.1016/j.jbi.2014.07.011. ISSN 1532-0464. PMID 25046831. PMC 4261011. http://dx.doi.org/10.1016/j.jbi.2014.07.011.
- ↑ Maver, Ales; Hristovski, Dimitar; Rindflesch, Thomas C.; Peterlin, Borut (2013-11-24). "Integration of Data from Omic Studies with the Literature-Based Discovery towards Identification of Novel Treatments for Neovascularization in Diabetic Retinopathy" (in en). BioMed Research International 2013: e848952. doi:10.1155/2013/848952. ISSN 2314-6133. PMID 24350292.
- ↑ Kostoff, Ronald N.; Briggs, Michael B. (February 2008). "Literature-Related Discovery (LRD): Potential treatments for Parkinson's Disease". Technological Forecasting and Social Change 75 (2): 226–238. doi:10.1016/j.techfore.2007.11.007. ISSN 0040-1625. http://dx.doi.org/10.1016/j.techfore.2007.11.007.
- ↑ Dong, Weiwei; Liu, Yixuan; Zhu, Weijie; Mou, Quan; Wang, Jinliang; Hu, Yi (2014-06-20). "Simulation of Swanson's literature-based discovery: anandamide treatment inhibits growth of gastric cancer cells in vitro and in silico" (in English). PLOS ONE 9 (6): e100436. doi:10.1371/JOURNAL.PONE.0100436. PMID 24949851. PMC 4065097. Bibcode: 2014PLoSO...9j0436D. https://www.wikidata.org/wiki/Q33784314.
- ↑ Kostoff, Ronald N.; Briggs, Michael B.; Lyons, Terence J. (February 2008). "Literature-related discovery (LRD): Potential treatments for Multiple Sclerosis". Technological Forecasting and Social Change 75 (2): 239–255. doi:10.1016/j.techfore.2007.11.002. ISSN 0040-1625. http://dx.doi.org/10.1016/j.techfore.2007.11.002.
- ↑ Hristovski, Dimitar; B, Peterlin; S, Dzeroski (2001-01-01). "Literature-based Discovery Support System and Its Application to Disease Gene Identification.". Proceedings. AMIA Annual Symposium: 928. PMC 2243305. https://www.wikidata.org/wiki/Q64946889.
- ↑ Sarkar, Indra Neil; Agrawal, Abha (2006). "Literature based discovery of gene clusters using phylogenetic methods". AMIA ... Annual Symposium Proceedings. AMIA Symposium 2006: 689–693. ISSN 1942-597X. PMID 17238429.
- ↑ Ahlers, Caroline B.; Hristovski, Dimitar; Kilicoglu, Halil; Rindflesch, Thomas C. (2007-10-11). "Using the literature-based discovery paradigm to investigate drug mechanisms". AMIA ... Annual Symposium Proceedings. AMIA Symposium 2007: 6–10. ISSN 1942-597X. PMID 18693787.
- ↑ Srinivasan, Mythily; Blackburn, Corinne; Mohamed, Mohamed; Sivagami, A. V.; Blum, Janice S. (2015-05-14). "Literature-based discovery of salivary biomarkers for type 2 diabetes mellitus" (in English). Biomarker Insights 10: 39–45. doi:10.4137/BMI.S22177. PMID 26005324. PMC 4433061. https://www.wikidata.org/wiki/Q35610345.
- ↑ Malec, Scott A.; Wei, Peng; Xu, Hua; Bernstam, Elmer V.; Myneni, Sahiti; Cohen, Trevor (2016-01-01). "Literature-Based Discovery of Confounding in Observational Clinical Data". AMIA Annual Symposium Proceedings 2016: 1920–1929. PMID 28269951. PMC 5333204. https://www.wikidata.org/wiki/Q31172385.
- ↑ Dai, Zhenguo; Li, Qian; Yang, Guang; Wang, Yini; Liu, Yang; Zheng, Zhilei; Tu, Yingfeng; Yang, Shuang et al. (2019-06-11). "Using literature-based discovery to identify candidate genes for the interaction between myocardial infarction and depression". BMC Medical Genetics 20 (1): 104. doi:10.1186/S12881-019-0841-8. PMID 31185929. PMC 6560897. https://www.wikidata.org/wiki/Q92663755.
- ↑ Vos, Rein; Aarts, Sil; Mulligen, Erik M. van; Metsemakers, Job; Boxtel, Martin P. van; Verhey, Frans RJ; Akker, Marjan van den (2013-06-17). "Finding potentially new multimorbidity patterns of psychiatric and somatic diseases: exploring the use of literature-based discovery in primary care research.". Journal of the American Medical Informatics Association 21 (1): 139–145. doi:10.1136/AMIAJNL-2012-001448. PMID 23775174. PMC 3912726. https://www.wikidata.org/wiki/Q37551215.
- ↑ Kostoff, Ronald N.; Solka, Jeffrey L.; Rushenberg, Robert L.; Wyatt, Jeffrey A. (February 2008). "Literature-related discovery (LRD): Water purification". Technological Forecasting and Social Change 75 (2): 256–275. doi:10.1016/j.techfore.2007.11.009. ISSN 0040-1625. http://dx.doi.org/10.1016/j.techfore.2007.11.009.
- ↑ Gordon, M. D.; Awad, N. F. (2008), "The Tip of the Iceberg: The Quest for Innovation at the Base of the Pyramid", Literature-based Discovery (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 23–37, doi:10.1007/978-3-540-68690-3_3, ISBN 978-3-540-68685-9, Bibcode: 2008lbd..book...23G, http://dx.doi.org/10.1007/978-3-540-68690-3_3, retrieved 2022-03-15
- ↑ Hristovski, Dimitar; Kastrin, Andrej; Rindflesch, Thomas C. (2015-08-25). "Semantics-Based Cross-domain Collaboration Recommendation in the Life Sciences". Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 2015. New York, NY, USA: ACM. pp. 805–806. doi:10.1145/2808797.2809300. ISBN 9781450338547. http://dx.doi.org/10.1145/2808797.2809300.
 |



