Biology:SKIDA1
 Generic protein structure example |
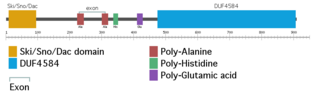
Ski/Dach domain-containing protein 1 is a protein that in humans is encoded by the SKIDA1 gene.[1] It is also known as C10orf140 and DLN-1. It has orthologs in vertebrates. It has two domains: the Ski/Sno/Dac domain and a domain of unknown function, DUF4854. It is associated with multiple types of cancer, like leukemia, ovarian cancer, and colon cancer.[2][3] It's predicted to be a nuclear protein.[4] It may interact with PRC2.[5][6]
Homologs
Orthologs
SKIDA1 has orthologs in vertebrate species. The species least related to humans with a SKIDA1 ortholog is the lancelet Branchiostoma belcheri. The clades amphibia and chondrichthyes have at least two species with SKIDA1, but SKIDA1 is not found throughout the clades. No orthologs have been found in lungfish or invertebrate species.[7]
Paralogous Domains
SKIDA1 shares the Ski/Sno/Dac domain with Ski oncogene (Ski), Ski-like protein (Sno), and dachshund (Dac).[8] It shares DUF4584 with Elongin BC Polycomb Repressive Complex 2 associated Protein (EPOP).[1]
Structure
In humans, SKIDA1 is located on the reverse strand of chromosome 10 at locus 10p12.31. It contains five exons.[1]
Isoforms
There is not a consensus on whether humans have one or two SKIDA1 isoforms. NCBI Gene claims there is one, while UniProt claims there are two.[9][10] It's possible isoform 2 is recorded in NCBI Gene as DLN-1 (accession BAE93016.1). Isoform 1 is 908 amino acids long, while isoform 2 is 827 amino acids long; isoform 2 is missing amino acids 240-318 from isoform 1.[10] Isoform 1 is predicted to weigh 98 kDa and have an isoelectric point of 8.7, while isoform 2 is predicted to weigh 90 kDa and have an isoelectric point of 7.6.[11]
Other mammalian species also have multiple isoforms of SKIDA1, including carnivorans, rodents, and primates. The number of isoforms each species has varies: cheetahs have five recorded isoforms, chimpanzees have three recorded, and brown rats have two recorded.[12]
Amino Acid Repeats
Human SKIDA1 contains two poly-alanine regions, one poly-histidine region, and one poly-glutamic acid region.[1] It's unknown if they have any function. The poly-alanine and poly-histidine regions are not highly conserved among orthologs; for example, while they are found in the house mouse ortholog, they are not found in the western lowland gorilla ortholog.[13][14] The poly-glutamic acid region shows more conservation, and is found abbreviated in species as distantly related from humans as the tire track eel.[15]
Domains
SKIDA1 contains two domains: Ski/Sno/Dac and DUF4854. The Ski/Sno/Dac domain is at the N-terminus end of the protein. The Ski/Sno/Dac domain is also found in the proteins Ski, Ski-like protein, and dachshund.[8] It is potentially a DNA-binding domain.[16]
The other domain, DUF4854, is also found in EPOP, near its C-terminus. However, the DUF4584 found in EPOP is roughly a fifth the size of that in SKIDA1. The C-termini of SKIDA1 (amino acids 844-908) and EPOP (amino acids 313-379) have 52% identity. The C-terminus of EPOP binds to the SUZ12 subunit of Polycomb Repressive Complex 2 (PRC2), suggesting that of SKIDA1 may as well.[5]
Regulation
Promoter and Transcription Factors
In humans, there are five predicted potential promoters. Two align with the second half of the mRNA transcript, suggesting they are not used or only produce an incomplete polypeptide.[17]
The promoter that aligns best with the start of the mRNA transcript is potentially bound to by many transcription factors, including Transcription factor II B, Nuclear factor Y, Early growth response 1, and Krueppel-like factor 6.[17] It does not contain a TATA box.
Transcript Regulation
SKIDA1 is regulated by microRNAs. miR-93 binds to the SKIDA1 3'-UTR.[18] Multiple microRNAs are predicted to bind to the SKIDA1 3'-UTR, including miR-130, miR-301, miR-454, and miR-494.[19]
Polypeptide Modification
SKIDA1 is SUMOylated at five sites.[20] Additional sites are predicted to be SUMOylated.[21][22] SKIDA1 is also predicted to be phosphorylated and O-GlcNAcylated.[23][24]
Expression
Subcellular Localization
SKIDA1 is predicted to be localized primarily in the nucleus and less so in the cytosol.[4]
Tissue Expression
SKIDA1 is expressed at high levels in the brain, thyroid, and testes. It's expressed at medium to low levels in adipose tissue, lymph nodes, and skeletal muscle.[25][26][27][28] In mice, it's noted to have medium-to-high expression in the olfactory bulb, retina, and salivary gland.[25]
Developmental Expression
SKIDA1 expression changes during organism development. Expression is low in the zygote, peaks during embryonic development, and is low post-birth. In the house mouse, it's expressed most during organogenesis.[29] In the fetus, its expression is low in the liver but not other organs.[30] Expression in the adult liver is much higher. In contrast, SKIDA1 expression in the fetal brain is higher than in the adult brain.[28]
SKIDA1 in the African clawed frog is expressed faintly in the marginal zone of gastrulae. During neurulation, it's expressed in the brain and cranial neural crest. During tailbud, SKIDA1 expression increases in sensory placodes. By the end of tailbud, neural expression has faded except in the olfactory organ.[31]
Function
SKIDA1 is predicted to function primarily in the nucleus and also in the cytosol.[4]
SKIDA1 knockouts in mice have significant differences from wild-type mice in the skeletal, neurological, reproductive, and immune systems. Other significant differences include effected hearing, an enlarged thymus, and increased pre-weaning mortality.[32] Some, but not all, of these effects were found in heterozygous knockouts.
Clinical significance
SKIDA1 expression is associated with multiple types of cancer. It is over-expressed in epithelial ovarian cancer cells.[33] Its expression is altered by various cancer-treatment compounds: human alpha-lactalbumin made lethal to tumor cells; oleate salts; metformin; and aspirin.[citation needed] In cell lines of cancerous cells, altered expression is associated with resistance to dasatinib and docetaxel, which are used to treat cancer.[34][35]
Altered methylation of SKIDA1 is associated with human pancreatic cancer, rheumatoid arthritis, and lupus erythematosus.[36][37] Additionally, SKIDA1 is expressed less in women with Down syndrome compared to their identical twins without Down syndrome.[38] Its expression is dramatically reduced in brains affected by untreated HIV1-associated neurocognitive disorders (HAND) in comparison to healthy brains and brains affected by HAND but treated with antiretrovirals.[39]
References
- ↑ 1.0 1.1 1.2 1.3 "SKI/DACH domain-containing protein 1 [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/protein/148762982.
- ↑ "GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer". Nature Genetics 45 (4): 362–70, 370e1-2. April 2013. doi:10.1038/ng.2564. PMID 23535730.
- ↑ "A molecular portrait of microsatellite instability across multiple cancers". Nature Communications 8: 15180. June 2017. doi:10.1038/ncomms15180. PMID 28585546. Bibcode: 2017NatCo...815180C.
- ↑ 4.0 4.1 4.2 "PSORT II". 24 November 1999. https://psort.hgc.jp/form2.html.
- ↑ 5.0 5.1 "The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells". Cell Discovery 1: 15008. 2015-04-28. doi:10.1038/celldisc.2015.8. PMID 27462409.
- ↑ "A High-Density Map for Navigating the Human Polycomb Complexome" (in English). Cell Reports 17 (2): 583–595. October 2016. doi:10.1016/j.celrep.2016.08.096. PMID 27705803.
- ↑ "NCBI Protein". https://www.ncbi.nlm.nih.gov/protein/.
- ↑ 8.0 8.1 "NCBI CDD CDD Conserved Protein Domain Ski_Sno". https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=308187.
- ↑ "SKIDA1 SKI/DACH domain containing 1 [Homo sapiens (human)"]. 13 February 2019. https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=387640.
- ↑ 10.0 10.1 "SKIDA1 - SKI/DACH domain-containing protein 1". 10 April 2019. https://www.uniprot.org/uniprot/Q1XH10.
- ↑ "Compute pI/Mw". https://web.expasy.org/compute_pi/.
- ↑ Protein [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004 – [cited 2019 March 2]. Available from: https://www.ncbi.nlm.nih.gov/protein/
- ↑ "SKI/DACH domain-containing protein 1 [Mus musculus"]. 15 August 2018. https://www.ncbi.nlm.nih.gov/protein/NP_082593.2.
- ↑ "PREDICTED: SKI/DACH domain-containing protein 1[Gorilla gorilla gorilla"]. 4 November 2016. https://www.ncbi.nlm.nih.gov/protein/XP_004049188.1.
- ↑ "SKI/Dach domain-containing protein 1 [Mastacembelus armatus"]. 6 September 2018. https://www.ncbi.nlm.nih.gov/protein/XP_026148649.1.
- ↑ "Structure of the retinal determination protein Dachshund reveals a DNA binding motif" (in English). Structure 10 (6): 787–95. June 2002. doi:10.1016/S0969-2126(02)00769-4. PMID 12057194.
- ↑ 17.0 17.1 "Genome Annotation and Browser". December 2017. https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl.
- ↑ Saito, Kazuyoshi (2011). "The mechanism of inflammation in autoimmune response during the acute phase of Kawasaki disease.". https://kaken.nii.ac.jp/en/report/KAKENHI-PROJECT-22790969/22790969seika/.
- ↑ "TargetScanHuman 7.2 predicted targeting of Human SKIDA1". March 2018. http://www.targetscan.org/cgi-bin/targetscan/vert_72/view_gene.cgi?rs=ENST00000449193.2&taxid=9606.
- ↑ "Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation". Nature Structural & Molecular Biology 24 (3): 325–336. March 2017. doi:10.1038/nsmb.3366. PMID 28112733.
- ↑ "SUMOplot". 2013. https://www.abgent.com/sumoplot.
- ↑ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". 2014. http://sumosp.biocuckoo.org/online.php.
- ↑ "GPS 3.0 - Kinase-specific Phosphorylation Site Prediction". http://gps.biocuckoo.org/online.php.
- ↑ "YinOYang 1.2 Server". 2 January 2017. http://www.cbs.dtu.dk/services/YinOYang.
- ↑ 25.0 25.1 "A gene atlas of the mouse and human protein-encoding transcriptomes". Proceedings of the National Academy of Sciences of the United States of America 101 (16): 6062–7. April 2004. doi:10.1073/pnas.0400782101. PMID 15075390. Bibcode: 2004PNAS..101.6062S.
- ↑ "Definition, conservation and epigenetics of housekeeping and tissue-enriched genes". BMC Genomics 10 (1): 269. June 2009. doi:10.1186/1471-2164-10-269. PMID 19534766.
- ↑ "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics". Molecular & Cellular Proteomics 13 (2): 397–406. February 2014. doi:10.1074/mcp.M113.035600. PMID 24309898.
- ↑ 28.0 28.1 "Genome-wide identification of zero nucleotide recursive splicing in Drosophila". Nature 521 (7552): 376–9. May 2015. doi:10.1038/nature14475. PMID 25970244. Bibcode: 2015Natur.521..376D.
- ↑ "EST Profile - Mm.102183". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.102183.
- ↑ "GenePaint". http://genepaint.org/results/skida1.
- ↑ "Expression of a novel Ski-like gene in Xenopus development". Gene Expression Patterns 6 (1): 22–8. December 2005. doi:10.1016/j.modgep.2005.05.004. PMID 16169285.
- ↑ "High-throughput discovery of novel developmental phenotypes". Nature 537 (7621): 508–514. September 2016. doi:10.1038/nature19356. PMID 27626380. Bibcode: 2016Natur.537..508..
- ↑ "GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer". Nature Genetics 45 (4): 362–70, 370e1-2. April 2013. doi:10.1038/ng.2564. PMID 23535730.
- ↑ "Activation of protein phosphatase 2A tumor suppressor as potential treatment of pancreatic cancer". Molecular Oncology 9 (4): 889–905. April 2015. doi:10.1016/j.molonc.2015.01.002. PMID 25637283.
- ↑ "Identification of docetaxel resistance genes in castration-resistant prostate cancer". Molecular Cancer Therapeutics 11 (2): 329–39. February 2012. doi:10.1158/1535-7163.MCT-11-0289. PMID 22027694.
- ↑ Ahlquist DA, Kisiel JB, Taylor WR, Yab TC, Mahoney DW, Lidgard GP, Allawi HT, "Detecting Neoplasm", US patent 9994911, issued 12 June 2018, assigned to Mayo Foundation for Medical Educationand Research Exact Sciences Development Company LLC
- ↑ "Epigenome-wide association study of rheumatoid arthritis identifies differentially methylated loci in B cells". Human Molecular Genetics 26 (14): 2803–2811. July 2017. doi:10.1093/hmg/ddx177. PMID 28475762.
- ↑ "Data in brief: Transcriptome analysis of induced pluripotent stem cells from monozygotic twins discordant for trisomy 21". Genomics Data 2: 226–9. December 2014. doi:10.1016/j.gdata.2014.07.006. PMID 26484098.
- ↑ Desrosiers, Ronald C., ed (September 2011). "Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders". PLOS Pathogens 7 (9): e1002213. doi:10.1371/journal.ppat.1002213. PMID 21909266.
 |