Biology:C4orf36
 Generic protein structure example |
C4orf36 (chromosome 4 open reading frame 36) is a protein that in humans is encoded by the c4orf36 gene.[1]
Gene
C4orf36 (also referred to as MGC26744, LOC132989, spostobu, fersnyby, and forsnyby)[2] is located on chromosome 4 at band 4q21.3 on the minus strand.[1] The gene is 21,052 base pairs long (chr4: 86,876,205-86,897,256)[3] and contains 8 exons.[1] Its gene neighborhood includes AFF1-AS1, RPL6P13, SLC10A6, LOC260422, and AFF1.[4] No human paralogs for c4orf36 have been identified.
Expression
RNA-seq and microarray data indicate that the c4orf36 gene is most highly expressed in the placenta, thyroid, ovary, and skin.[1][5][6] It is also expressed in the lungs during fetal development, most highly at 17 weeks into development.[1]
Transcript
C4orf36 encodes 24 different mRNA transcripts, including 20 alternatively spliced variants and 4 unspliced forms.[2] The transcript variant 1 (NM_144645.4), the subject of this article, is 908 nucleotides long and contains 5 exons.[7] The promoter for transcript variant 1 (GXP_263623) spans the base pairs 86892213-86893422 on chromosome 4.[8]
Protein
C4orf36 encodes a 117 amino acid protein[9] with a molecular weight of 13.28 kDa and an isoelectric point of 9.54.[10] It contains a divalent cation tolerance protein CutA motif from amino acids 57-88.[11] No transmembrane domains or N-terminal signal peptides have been identified for c4orf36.[12] The protein is predicted to undergo several post-translational modifications, including myristoylation and phosphorylation.[13][14]
Immunohistochemistry experiments have shown that the c4orf36 protein is localized to the cytosol and focal adhesion sites in the human U2-OS cell line, cultivated from the bone tissue of an osteosarcoma patient.[15][16] Staining in human kidney tissue showed strong cytoplasmic and membranous expression within cells of tubules and moderate expression in glomeruli.[17] Data from Human Protein Atlas also indicates that the protein is enriched in several regions of the brain, including the basal ganglia, cerebral cortex, pons, medulla, and in the hippocampus during its formation.[18]
Evolutionary history
C4orf36 orthologs are found in mammals and turtles. The gene is not found in other reptiles, birds, amphibians, fish, invertebrates, or outside of Kingdom Animalia. The gene likely first appeared around 318 million years ago, when mammals diverged from birds and reptiles.
The following table presents a diverse subset of c4orf36 orthologs found using BLAST searches.[19] Within mammals, the sequence identity of the selected orthologs ranges from 99.1% to 47%. Within turtles, the sequence identity ranges from 34.8% to 32.2%.
| Scientific Name | Common Name | NCBI Accession # | Protein Length (AA) | Sequence Identity to Humans (%) |
| Homo sapiens | Human | NP_653246.2 | 117 | 100.0 |
| Pan troglodytes | Chimpanzee | PNI41451.1 | 113 | 99.1 |
| Halichoerus grypus | Gray Seal | XP_035952662.1 | 117 | 75.2 |
| Equus caballus | Horse | XP_023493530 | 117 | 75.2 |
| Tursiops truncates | Common bottlenose dolphin | XP_019782327.1 | 117 | 75.2 |
| Manis pentadactyla | Chinese pangolin | XP_036752164.1 | 117 | 73.5 |
| Choloepus didactylus | Southern two-toed sloth | XP_037684921.1 | 117 | 73.5 |
| Galeopterus variegatus | Sunda flying lemur | XP_008566526.1 | 117 | 72.7 |
| Bos Taurus | Cattle | XP_015327194.1 | 117 | 72.7 |
| Vulpes Vulpes | Red fox | XP_025853915.1 | 117 | 68.4 |
| Condylura cristata | Star-nosed mole | XP_004681481.1 | 117 | 67.5 |
| Eptesicus fuscus | Big brown bat | XP_008141802.1 | 116 | 66.7 |
| Mus musculus | Mouse | NP_001157022 | 117 | 64.1 |
| Monodelphis domestica | Gray short-tailed opossum | XP_007495994 | 118 | 52.1 |
| Ornithorhynchus anatinus | Platypus | XP_007658949.1 | 159 | 47.0 |
| Trachemys scripta elegans | Slider turtle | XP_034628022.1 | 117 | 34.8 |
| Chrysemys picta bellii | Painted turtle | XP_042712362 | 117 | 34.8 |
| Dermochelys coriacea | Leatherback sea turtle | XP_038255787.1 | 117 | 33.9 |
| Chelonia mydas | Green sea turtle | XP_043401238.1 | 117 | 33.0 |
| Mauremys mutica | Yellow pond turtle | KAH1170002 | 117 | 32.2 |
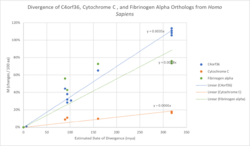
C4orf36 is predicted to evolve more rapidly than other common human proteins, including cytochrome C and fibrinogen alpha.
Clinical significance
Immunostaining of c4orf36 in syncytiotrophoblast cells (STBs) of mid-gestation human embryos showed a dense punctate pattern consistent with that of c4orf36’s fungal protein analog, dynein light chain, which is involved in intracellular cargo transport and other cellular processes.[20] In another study, ovarian cancer (OVCA) cell lines were treated with gemcitabine. Higher c4orf36 expression was associated with in vitro chemoresistance to the drug, indicating that increased c4orf36 expression may be related to decreased OCVA patient survival.[21] Furthermore, microarray data suggests that C4orf36 expression levels are higher in the saliva of pre-treatment pancreatic cancer patients than that of healthy controls.[22]
In one study on adaptogenic plants, neuroglia cell lines were treated with Rhaponticum carthamoides (RC), which promotes non-specific stress resistance and may mitigate the onset of senescence. C4orf36 was significantly deregulated in neuroglia cell lines after RC treatment, indicating that tighter c4orf36 regulation may play a role in the development of age-related disorders.[23] A study on chondrogenesis found that c4orf36 has an oscillatory expression pattern coupled to that of ATP, indicating a potential link to skeletal development during embryogenesis.[24]
Mutations
Chemically significant amino acid changes in conserved regions of the c4orf36 protein were found with NCBI SNPGeneView.[25]
| dbSNP rs# Cluster ID | Mutation | Function |
| rs149531474 | M1T | No protein |
| rs1200223723 | P70L | Missense |
| rs201168053 | E78* | Nonsense |
| rs908052987 | L86P | Missense |
| rs762808044 | P113R | Missense |
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "C4orf36 chromosome 4 open reading frame 36 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/132989.
- ↑ 2.0 2.1 "AceView: Gene:C4orf36, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=C4orf36.
- ↑ "Homo sapiens chromosome 4, GRCh38.p13 Primary Assembly" (in en-US). 2021-11-22. http://www.ncbi.nlm.nih.gov/nuccore/NC_000004.12.
- ↑ "Gene neighbors for Gene (Select 132989) - Gene - NCBI". https://www.ncbi.nlm.nih.gov/gene?LinkName=gene_gene_neighbors&from_uid=132989.
- ↑ "48992692 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/48992692.
- ↑ "69554904 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/?term=c4orf36,+GDS3834.
- ↑ "Homo sapiens chromosome 4 open reading frame 36 (C4orf36), transcript variant 1, mRNA" (in en-US). 2020-12-12. http://www.ncbi.nlm.nih.gov/nuccore/NM_144645.4.
- ↑ "Genomatix: ElDorado". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=a691e18792b81bb3a3ac1430949e9770;RESULT=C4orf36_1.
- ↑ "uncharacterized protein C4orf36 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/1519312833.
- ↑ "ExPASy: get PI/Mw". https://www.expasy.org/resources/compute-pi-mw.
- ↑ "GenomeNet MOTIF Search". https://www.genome.jp/tools/motif/.
- ↑ "PSORTII Prediction". https://psort.hgc.jp/form2.html.
- ↑ "ScanProsite". https://prosite.expasy.org/scanprosite/.
- ↑ "MyHits - SIB Swiss Institute of Bioinformatics | Expasy". https://www.expasy.org/resources/myhits.
- ↑ "C4orf36 Antibody (NBP2-31678): Novus Biologicals". https://www.novusbio.com/products/c4orf36-antibody_nbp2-31678#datasheet.
- ↑ "U-2 OS | ATCC". https://www.atcc.org/products/htb-96.
- ↑ "C4orf36 Antibody (PA5-63441)". https://www.thermofisher.com/antibody/product/C4orf36-Antibody-Polyclonal/PA5-63441.
- ↑ "C4orf36 protein expression summary - The Human Protein Atlas". https://www.proteinatlas.org/ENSG00000163633-C4orf36.
- ↑ "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "RNA profiling of laser microdissected human trophoblast subtypes at mid-gestation reveals a role for cannabinoid signaling in invasion". Development 148 (20): dev199626. October 2021. doi:10.1242/dev.199626. PMID 34557907.
- ↑ "The O-glycan pathway is associated with in vitro sensitivity to gemcitabine and overall survival from ovarian cancer". International Journal of Oncology 41 (1): 179–188. July 2012. doi:10.3892/ijo.2012.1451. PMID 22552627.
- ↑ "GDS4100 / 1552919_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS4100:1552919_at.
- ↑ "Synergy assessments of plant extracts used in the treatment of stress and aging-related disorders" (in en). Synergy 7: 39–48. 2018-12-01. doi:10.1016/j.synres.2018.10.001. ISSN 2213-7130.
- ↑ "Analysis of proteins showing differential changes during ATP oscillations in chondrogenesis". Cell Biochemistry and Function 32 (5): 429–437. July 2014. doi:10.1002/cbf.3033. PMID 24578328.
- ↑ "SNP linked to Gene (geneID:132989) Via Contig Annotation". https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=132989.
 |