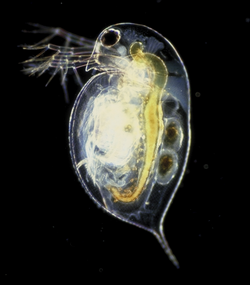
Biology:Daphnia pulex
| Daphnia pulex | |
|---|---|
| |
| Scientific classification Error creating thumbnail: Unable to save thumbnail to destination
| |
| Domain: | Eukaryota |
| Kingdom: | Animalia |
| Phylum: | Arthropoda |
| Class: | Branchiopoda |
| Order: | Anomopoda |
| Family: | Daphniidae |
| Genus: | Daphnia |
| Subgenus: | Daphnia |
| Species: | D. pulex
|
| Binomial name | |
| Daphnia pulex | |
Daphnia pulex is the most common species of water flea.[3] It has a cosmopolitan distribution: the species is found throughout the Americas, Europe, and Australia.[4] It is a model species, and was the first crustacean to have its genome sequenced.
Description
D. pulex is an arthropod whose body segments are difficult to distinguish. It can only be recognised by its appendages (only ever one pair per segment), and by studying its internal anatomy.[5] The head is distinct and is made up of six segments, which are fused together even as an embryo. It bears the mouthparts, and two pairs of antennae, the second pair of which is enlarged into powerful organs used for swimming.[5] No clear division is seen between the thorax and abdomen, which collectively bear five pairs of appendages.[5] The shell surrounding the animal extends posteriorly into a spine.[6] Like most other Daphnia species, D. pulex reproduces by cyclical parthenogenesis, alternating between sexual and asexual reproduction.[7]
Ecology
D. pulex occurs in a wide range of aquatic habitats, although it is most closely associated with small, shaded pools.[8] In oligotrophic lakes, D. pulex has little pigmentation, while it may become bright red in hypereutrophic waters, due to the production of haemoglobin.[8]
Predation
Daphnia species are prey for a variety of both vertebrate and invertebrate predators. The role of predation on D. pulex population ecology is extensively studied, and has been shown to be a major axis of variation in shaping population dynamics[9] and landscape-level distribution.[10] In addition to the direct population ecological effects of predation, the process contributes to phenotypic evolution in contrasting ways; larger D. pulex individuals are more visible to vertebrate predators, but invertebrate predators are unable to handle larger ones. As a result, larger water fleas tend to be found with invertebrate predators, while smaller size is associated with vertebrate predators.
Similar to some other Daphnia species, the morphology of D. pulex exhibits a plastic response to the presence of predators. Phantom midge larvae (Chaoborus) release kairomones – chemical cues – that induce the development of small, jagged protrusions on the head, known as "neck teeth",[11] which increase survivorship in the presence of the invertebrate predator, but at a cost – longer development time, for example – when those predators are not present.[12]
Ecological stoichiometry
D. pulex ecology is shaped by nutrient availability and balance, which affects traits that mediate intra- and interspecific interactions. Because nutrients are required for an array of biological processes – for example, amino acid synthesis – the environmental availability of these nutrients regulates downstream organismal characteristics.[13] Low nutrient availability reduces both body size and growth rate, which, as noted above, regulates Daphnia relationships to predators. D. pulex in particular has been an important model species for investigating ecological stoichiometry, demonstrating that pond shading by trees increases nutrient concentrations relative to carbon in algae, which increases D. pulex body size, and therefore competitive ability and susceptibility to predation by vertebrates.[14]
Genomics
D. pulex was the first crustacean to have its genome sequenced.[15][16] Its genome contains 31,000 genes – 8,000 more than are present in the human genome – as a result of extensive gene duplication.[17]
One of the most astonishing features of the D. pulex genome is its compactness: despite being around 200 megabase pairs (Mbp) in size (around 1/16th of that of the human genome, which is 3,200 Mbp in size); its 12 chromosomes contain a minimum set of 30,907 predicted protein-coding genes, more than the 20,000–25,000 contained in the human counterpart.[17]
Such an efficient gene packaging is achieved by means of a small intron size. Indeed, whereas the mean protein length in D. pulex is quite similar to that of Drosophila, the average gene size is 1000 bp shorter in D. pulex. As inferred from expressed sequence tag analysis, the average intron size found in D. pulex genes is 170 bp.[17]
The intron density of the D. pulex genome, though, is similar to that of Apis mellifera, which in turn is twice that found in Drosophila.[17]
The D. pulex genome has undergone extensive gene duplication followed by rapid paralog divergence and tandem rearrangement. As a result of these processes, around 20% of its gene catalog is composed of tandems consisting of three to 80 paralog genes, many of which are ecoresponsive, that is, they are expressed differently upon exposure of D. pulex to environmental challenges such as biotic or abiotic stress or fluctuations in light or oxygen levels.[17]
Parthenogenesis
D. pulex can reproduce by cyclical parthenogenesis or obligate parthenogenesis.[18] During cyclical parthenogenesis D. pulex cycles between a sexual stage and a parthenogenetic stage. During the sexual stage females produce haploid eggs by meiosis, and these eggs require fertilization by a male to develop further. During the parthenogenetic stage, eggs are produced that do not require fertilization to develop further. The lineages of D. pulex that reproduce by obligate parthenogenesis also do not require fertilization and produce direct-developing eggs that are indistinguishable from eggs produced by parthenogenesis in cyclical lineages.[18] Parthenogenesis appears to involve initial meiotic chromosome pairing. During both cyclic and obligate parthenogenesis a polar body is extruded during cell division indicating initiation of meiosis.
Notes
References
- ↑ Gregorio Fernandez-Leborans; Maria Luisa Tato-Porto (2000). "A review of the species of protozoan epibionts on crustaceans. II. Suctorian ciliates". Crustaceana 73 (10): 1205–1237. doi:10.1163/156854000505209.
- ↑ "Daphnia pulex Leydig, 1860". Integrated Taxonomic Information System. https://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=83874.
- ↑ Carrie Miller. "Daphnia pulex". Animal Diversity Web. University of Michigan. http://animaldiversity.ummz.umich.edu/site/accounts/information/Daphnia_pulex.html.
- ↑ "Daphnia pulex". An Image-Based Key To The Zooplankton Of The Northeast (USA). University of New Hampshire. http://cfb.unh.edu/CFBkey/html/Organisms/CCladocera/FDaphnidae/GDaphnia/Daphnia_pulex/daphniapulex.html.
- ↑ 5.0 5.1 5.2 Alexander Ivanovitch Petrunkevitch (1916). "Daphnia pulex". Morphology of Invertebrate Types. BiblioBazaar. pp. 113–121. ISBN 978-0-554-71763-0. https://books.google.com/books?id=w2hFvQik9VEC&pg=PA113.
- ↑ Herrick, Clarence Luther (2009). "Section 6". A Final Report on the Crustacea of Minnesota. General Books LLC. pp. 21–66. ISBN 978-1-150-02333-0. https://books.google.com/books?id=6lMD45YD94cC&pg=PA49.
- ↑ Eads, BD; Bohuski, E; Andrews, J (18 Dec 2007). "Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex". BMC Genomics 8 (2007): 464. doi:10.1186/1471-2164-8-464. PMID 18088424.
- ↑ 8.0 8.1 "Daphnia pulex". An Image-Based Key To The Zooplankton Of The Northeast (USA). Version 4.0. University of New Hampshire. http://cfb.unh.edu/cfbkey/html/organisms/ccladocera/fdaphnidae/gdaphnia/daphnia_pulex/daphniapulex.html.
- ↑ Barbara Leoni; Letizia Garibaldi (2009). "Population dynamics of Chaoborus flavicans and Daphnia spp.: effects on a zooplankton community in a volcanic eutrophic lake with naturally high metal concentrations (L. Monticchio Grande, Southern Italy)". Journal of Limnology 68 (1): 37–45. doi:10.4081/jlimnol.2009.37. https://jlimnol.it/index.php/jlimnol/article/view/jlimnol.2009.37/155.
- ↑ J. H. Pantel; T. E. Juenger; M. A. Leibold (2011). "Environmental gradients structure Daphnia pulex × pulicaria clonal distribution". Journal of Evolutionary Biology 24 (4): 723–732. doi:10.1111/j.1420-9101.2010.02196.x. PMID 21288271.
- ↑ Winfried Lampert; Ulrich Sommer (2007). "Predation". Limnoecology: The Ecology of Lakes and Streams (2nd ed.). Oxford University Press. pp. 162–179. ISBN 978-0-19-921393-1. https://books.google.com/books?id=zEcCX5k3hJAC&pg=PA176.
- ↑ R. Tollrian (1993). "Neckteeth formation in Daphnia pulex as an example of continuous phenotypic plasticity: morphological effects of Chaoborus kairomone concentration and their quantification". Journal of Plankton Research 15 (11): 1309–1318. doi:10.1093/plankt/15.11.1309.
- ↑ Robert Warner Sterner; James J. Elser (2002). Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere. Princeton University Press. ISBN 978-0-691-07491-7.
- ↑ Spencer R. Hall; Mathew A. Leibold; David A. Lytle; Val H. Smith (2004). "Stoichiometry and planktonic grazer composition over gradients of light, nutrients, and predation risk". Ecology 85 (8): 2291–2301. doi:10.1890/03-0471. Bibcode: 2004Ecol...85.2291H.
- ↑ "Daphnia pulex v1.0". DOE Joint Genome Institute. http://genome.jgi-psf.org/Dappu1/Dappu1.home.html.
- ↑ Florian Odronitz; Sebastian Becker; Martin Kollmar (2009). "Reconstructing the phylogeny of 21 completely sequenced arthropod species based on their motor proteins". BMC Genomics 10: 173. doi:10.1186/1471-2164-10-173. PMID 19383156.
- ↑ 17.0 17.1 17.2 17.3 17.4 John K. Colbourne et al. (2011). "The ecoresponsive genome of Daphnia pulex". Science 331 (6017): 555–561. doi:10.1126/science.1197761. PMID 21292972. Bibcode: 2011Sci...331..555C.
- ↑ 18.0 18.1 Schurko, A. M.; Logsdon Jr, J. M.; Eads, B. D. (2009). "Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution". BMC Evolutionary Biology 9 (1): 78. doi:10.1186/1471-2148-9-78. PMID 19383157. Bibcode: 2009BMCEE...9...78S.
Wikidata ☰ Q1080188 entry
 |

