Biology:Isochorismate lyase
| Isochorismate pyruvate lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
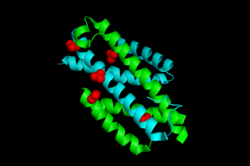 Native Isochorismate pyruvate lyase cartoon view with NO3 | |||||||||
| Identifiers | |||||||||
| EC number | 4.2.99.21 | ||||||||
| CAS number | 383896-77-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
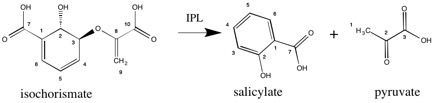
Isochorismate pyruvate lyase (IPL, EC 4.2.99.21) is an enzyme responsible for catalyzing part of the pathway involved in the formation of salicylic acid. More specifically, IPL will use isochorismate as a substrate and convert it into salicylate and pyruvate. IPL is a PchB enzyme originating from the pchB gene in Pseudomonas aeruginosa.[1]
Nomenclature
Isochorismate lyase, or PchB, is part of the lyase class of enzymes. These enzymes are typically responsible for the addition and removal of functional groups from substrates. The systematic name of this enzyme is isochorismate pyruvate-lyase (salicylate-forming). As its name implies, the substrate of this lyase is isochorismate. Derived from the pchB gene of Pseudomonas aeruginosa, other names for isochorismate lyase include:[2]
- salicylate biosynthesis protein PchB
- pyochelin biosynthetic protein PchB
- isochorismate pyruvate lyase
- PchB
- IPL.
Reaction
Isochorismate lyase's only substrate is isochorismate. It catalyzes the elimination of the enolpyruvyl side chain from isochorismate to make salicylate, a precursor to the siderophore pyochelin.[2] Believed to be a pericyclic reaction, the enzyme's transition state, when transferring a hydrogen from C2 to C9,[3] is cyclic. Its bond breaking and subsequent formation are conjoint processes. Note, however, that while conjoint, the breaking and forming of bonds will not necessarily occur at the same rate.[4] This enzyme functions at an optimal temperature of 25 °C and a pH of 6.8 to 7.5, but it is active across the entire pH range of 4-9. The Pseudomonas aeruginosa species does not require any cofactors or metals to complete the reaction.[2]
Structure
IPL consists of an intertwined dimer with two equal active sites. Each monomer has three helices, and both actives sites are composed of residues from each of the two monomers.[4] A loop between the first and second helices provide an opening for the substrate to enter and product to exit.[5] In that loop is a positively charged lysine at residue 42, which acts as a lid for this active site. This lysine is essential for the enzyme's most efficient method of producing salicylate. It is the positive charge on lysine 42, combined with the organization of the substrate once it enters the active site, that allows for the enzyme's transition state stabilization.[3] Before being bound to its substrate, IPL will have nitrates bound to its open active site.[5]
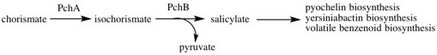
Pathway
Isochorismate lyase is the second enzyme in the pathway salicylate biosynthesis I, a pathway occurring immediately following chorismate biosynthesis I. Using the chorismate produced from that metabolic process, first the enzyme PchA will catalyze the reaction of chorismate into isochorismate, which in turn is used by IPL to cleave off a pyruvate, leaving the product salicylate.[6]
Salicylate
Salicylate is an aromatic compound found in species from both the plant and bacterial kingdoms. Its functions vary depending on the species, but generally salicylate can be used in many bacteria as a building block for various siderophores (organic Fe3+ chelators). These siderophores vary per organism. Some examples include yersiniabactin in the organisms Yersinia pestis and Yersinia enterocolitica, mycobactin in Mycobacterium tuberculosis, and pyochelin in Pseudomonas aeruginosa.[6]
Salicylate can also be found in various species of plants, where it is used for flowering and resistance against pathogen infections.[6]
Homologs
Isochorismate lyase is a structural homolog of the chorismate mutase enzyme in E. coli, and actually exhibits non-physiological chorismate mutase activity, albeit at a much lower efficiency.[3]
IPL also has several homologs found in other organisms, including:[2]
- Irp9 in Yersinia enterocolitica
- Mbtl or salicylate synthase in Mycobacterium tuberculosis
References
- ↑ "Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities". The Journal of Biological Chemistry 277 (24): 21768–75. June 2002. doi:10.1074/jbc.M202410200. PMID 11937513. http://www.jbc.org/content/277/24/21768.long. Retrieved April 2, 2015.
- ↑ 2.0 2.1 2.2 2.3 "BRENDA - Information on EC 4.2.99.21 - isochorismate lyase and Organism(s) Pseudomonas aeruginosa". http://www.brenda-enzymes.info/enzyme.php?ecno=4.2.99.21&Suchword=&organism%5B%5D=Pseudomonas+aeruginosa&show_tm=0.
- ↑ 3.0 3.1 3.2 "Modification of residue 42 of the active site loop with a lysine-mimetic side chain rescues isochorismate-pyruvate lyase activity in Pseudomonas aeruginosa PchB". Biochemistry 51 (38): 7525–32. September 2012. doi:10.1021/bi300472n. PMID 22970849.
- ↑ 4.0 4.1 "pH Dependence of catalysis by Pseudomonas aeruginosa isochorismate-pyruvate lyase: implications for transition state stabilization and the role of lysine 42". Biochemistry 50 (33): 7198–207. August 2011. doi:10.1021/bi200599j. PMID 21751784.
- ↑ 5.0 5.1 "Two crystal structures of the isochorismate pyruvate lyase from Pseudomonas aeruginosa". The Journal of Biological Chemistry 281 (44): 33441–9. November 2006. doi:10.1074/jbc.M605470200. PMID 16914555.
- ↑ 6.0 6.1 6.2 Caspi, R (December 18, 2009). "MetaCyc Pathway: salicylate biosynthesis I". SRI International. http://biocyc.org/META/new-image?type=PATHWAY&object=PWY-6406&detail-level=2&ENZORG=TAX-287.
 |