Biology:Tryptophan synthase
| Tryptophan Synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
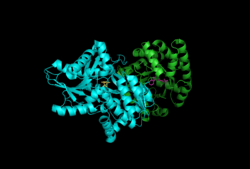 Subunits: Beta Subunit, Alpha Subunit with PLP, IGP | |||||||||
| Identifiers | |||||||||
| EC number | 4.2.1.20 | ||||||||
| CAS number | 9014-52-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Tryptophan synthase or tryptophan synthetase is an enzyme (EC 4.2.1.20) that catalyses the final two steps in the biosynthesis of tryptophan.[1][2] It is commonly found in Eubacteria,[3] Archaebacteria,[4] Protista,[5] Fungi,[6] and Plantae.[7] However, it is absent from Animalia.[8] It is typically found as an α2β2 tetramer.[9][10] The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 angstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling.[11] The active sites of tryptophan synthase are allosterically coupled.[12]
Enzyme structure
Subunits: Tryptophan synthase typically exists as an α-ββ-α complex. The α and β subunits have molecular masses of 27 and 43 kDa respectively. The α subunit has a TIM barrel conformation. The β subunit has a fold type II conformation and a binding site adjacent to the active site for monovalent cations.[13] Their assembly into a complex leads to structural changes in both subunits resulting in reciprocal activation. There are two main mechanisms for intersubunit communication. First, the COMM domain of the β-subunit and the α-loop2 of the α-subunit interact. Additionally, there are interactions between the αGly181 and βSer178 residues.[14] The active sites are regulated allosterically and undergo transitions between open, inactive, and closed, active, states.[12]
Indole-3-glycerol binding site: See image 1.
Indole and serine binding site: See image 1.
Hydrophobic channel: The α and β active sites are separated by a 25 angstrom long hydrophobic channel contained within the enzyme allowing for the diffusion of indole. If the channel did not exist, the indole formed at an α active site would quickly diffuse away and be lost to the cell as it is hydrophobic and can easily cross membranes. As such, the channel is essential for enzyme complex function.[15]
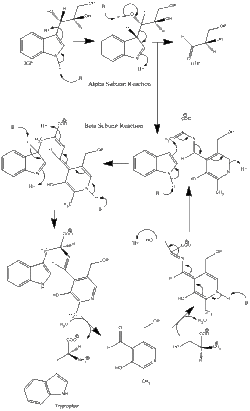
Enzyme mechanism
α subunit reaction: The α subunit catalyzes the formation of indole and G3P from a retro-aldol cleavage of IGP. The αGlu49 and αAsp60 are thought to be directly involved in the catalysis as shown.[11] The rate limiting step is the isomerization of IGP.[16] See image 2.
β subunit reaction: The β subunit catalyzes the β-replacement reaction in which indole and serine condense to form tryptophan in a PLP dependent reaction. The βLys87, βGlu109, and βSer377 are thought to be directly involved in the catalysis as shown.[11] Again, the exact mechanism has not been conclusively determined. See image 2.
Net reaction: See image 3.
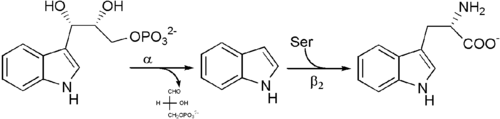
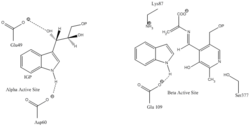
Biological function
Tryptophan synthase is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. It is absent from animals such as humans. Tryptophan is one of the twenty standard amino acids and one of nine essential amino acids for humans. As such, tryptophan is a necessary component of the human diet.
Substrate scope
Tryptophan synthetase is also known to accept indole analogues, e.g., fluorinated or methylated indoles, as substrates, generating the corresponding tryptophan analogues.[17]
Disease relevance
As humans do not have tryptophan synthase, this enzyme has been explored as a potential drug target.[18] However, it is thought that bacteria have alternate mechanisms to produce amino acids which might make this approach less effective. In either case, even if the drug only weakens bacteria, it might still be useful as the bacteria are already vulnerable in the hostile host environment. As such, the inhibition of tryptophan synthase along with other PLP-enzymes in amino acid metabolism has the potential to help solve medical problems.[19]
Inhibition of tryptophan synthase and other PLP-enzymes in amino acid metabolism has been suggested for:
- Treatment of tuberculosis[18]
- Treatment of ocular and genital infections[20]
- Treatment of cryptosporidiosis[18]
- Herbicide use[21]
Evolution
It is thought that early in evolution the trpB2 gene was duplicated. One copy entered the trp operon as trpB2i allowing for its expression with trpA. TrpB2i formed transient complexes with TrpA and in the process activated TrpA unidirectionally. The other copy remained outside as trpB2o, and fulfilled an existing role or played a new one such as acting as a salvage protein for indole. TrpB2i evolved into TrpB1, which formed permanent complexes with trpA resulting in bidirectional activation. The advantage of the indole salvage protein declined and the TrpB gene was lost. Finally, the TrpB1 and TrpA genes were fused resulting in the formation the bifunctional enzyme.[22]
Historical significance
Tryptophan synthase was the first enzyme identified that had two catalytic capabilities that were extensively studied. It was also the first identified to utilize substrate channeling. As such, this enzyme has been studied extensively and is the subject of great interest.[11]
See also
References
- ↑ "Tryptophan synthase: the workings of a channeling nanomachine". Trends in Biochemical Sciences 33 (6): 254–64. June 2008. doi:10.1016/j.tibs.2008.04.008. PMID 18486479.
- ↑ "Structural basis for catalysis by tryptophan synthase". Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology and Related Areas of Molecular Biology. 64. 1991. pp. 93–172. doi:10.1002/9780470123102.ch3. ISBN 9780470123102.
- ↑ "Tryptophan synthase: Identification of Pasteurella multocida tryptophan synthase B-subunit by antisera against strain PI059". Microbiology 142: 115–21. September 1996. doi:10.1099/13500872-142-1-115. PMID 8581158.
- ↑ "On the levels of enzymatic substrate specificity: Implications for the early evolution of metabolic pathways". Advances in Space Research 15 (3): 345–56. March 1995. doi:10.1016/S0273-1177(99)80106-9. PMID 11539248.
- ↑ "Gene Discovery in the Acanthamoeba castellanii Genome". Protist 156 (2): 203–14. August 2005. doi:10.1016/j.protis.2005.04.001. PMID 16171187.
- ↑ "The tryptophan synthetase gene TRP1 of Nodulisporium sp.: molecular characterization and its relation to nodulisporic acid A production". Appl Microbiol Biotechnol 79 (3): 451–9. April 2008. doi:10.1007/s00253-008-1440-3. PMID 18389234.
- ↑ Sanjaya, Hsiao PY, Su RC, Ko SS, Tong CG, Yang RY, Chan MT (April 2008). "Overexpression of Arabidopsis thaliana tryptophan synthase beta 1 (AtTSB1) in Arabidopsis and tomato confers tolerance to cadmium stress". Plant Cell Environ 31 (8): 1074–85. doi:10.1111/j.1365-3040.2008.01819.x. PMID 18419734.
- ↑ "The tryptophan synthase-encoding trpB gene of Aspergillus nidulans is regulated by the cross-pathway control system". Mol Gen Genet 263 (5): 867–76. June 2000. doi:10.1007/s004380000250. PMID 10905354.
- ↑ "Crystallization and preliminary X-ray crystallographic data of the tryptophan synthase alpha 2 beta 2 complex from Salmonella typhimurium". The Journal of Biological Chemistry 260 (6): 3716–3718. March 1985. doi:10.1016/s0021-9258(19)83682-7. PMID 3882715.
- ↑ "Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium". The Journal of Biological Chemistry 263 (33): 17857–17871. November 1988. doi:10.1016/s0021-9258(19)77913-7. PMID 3053720.
- ↑ 11.0 11.1 11.2 11.3 "Tryptophan synthase: a mine for enzymologists". Cell Mol Life Sci 66 (14): 2391–403. April 2009. doi:10.1007/s00018-009-0028-0. PMID 19387555.
- ↑ 12.0 12.1 "Synergistic regulation and ligand-induced conformational changes of tryptophan synthase". Biochemistry 48 (41): 9921–31. September 2009. doi:10.1021/bi901358j. PMID 19764814.
- ↑ "Modeling of the spatial structure of ornithine decarboxylases". Protein Sci 4 (7): 1291–304. July 1995. doi:10.1002/pro.5560040705. PMID 7670372.
- ↑ "Loop closure and intersubunit communication in tryptophan synthase". Biochemistry 37 (16): 5394–406. April 1998. doi:10.1021/bi9728957. PMID 9548921.
- ↑ "Channeling of Substrates and Intermediates in Enzyme-Catalyzes Reactions". Annu Rev Biochem 70: 149–80. 2001. doi:10.1146/annurev.biochem.70.1.149. PMID 11395405.
- ↑ "Serine modulates substrate channeling in tryptophan synthase. A novel intersubunit triggering mechanism". J Biol Chem 266 (13): 8020–33. May 1991. doi:10.1016/S0021-9258(18)92934-0. PMID 1902468.
- ↑ "The enzymatic synthesis of L-tryptophan analogues". Analytical Biochemistry 59 (2): 436–440. June 1974. doi:10.1016/0003-2697(74)90296-6. PMID 4600987.
- ↑ 18.0 18.1 18.2 "Protozoan genomics for drug discovery". Nat Biotechnol 23 (9): 1089–91. September 2005. doi:10.1038/nbt0905-1089. PMID 16151400.
- ↑ "Robust Salmonella metabolism limits possibilities for new antimicrobials". Nature 440 (7082): 303–7. March 2006. doi:10.1038/nature04616. PMID 16541065.
- ↑ "Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates". J Clin Invest 111 (11): 1757–69. June 2003. doi:10.1172/JCI17993. PMID 12782678.
- ↑ "On the structural basis of the catalytic mechanism and the regulation of the α-subunit of tryptophan synthase from Salmonella typhimurium and BXI from maize, two evolutionarily related enzymes". J Mol Biol 352 (3): 608–20. September 2005. doi:10.1016/j.jmb.2005.07.014. PMID 16120446.
- ↑ "Evolution of Multi-Enzyme Complexes: The Case of Tryptophan Synthase". Biochemistry 45 (47): 14111–9. November 2006. doi:10.1021/bi061684b. PMID 17115706.
 |
