Biology:BglII

| Restriction endonuclease BglII | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of restriction endonuclease BstYI bound to non-cognate DNA | |||||||||
| Identifiers | |||||||||
| Symbol | Endonuc-BglII | ||||||||
| Pfam | PF09195 | ||||||||
| Pfam clan | CL0236 | ||||||||
| InterPro | IPR015278 | ||||||||
| SCOP2 | 1dfm / SCOPe / SUPFAM | ||||||||
| |||||||||
BglII is a type II restriction endonuclease isolated from certain strains of Bacillus globigii.
The principal function of restriction enzymes is the protection of the host genome against foreign DNA, but they may also have some involvement in recombination and transposition.[1]
Like most type II restriction enzymes, BglII consists of two identical subunits that form a homodimer around the DNA double helix. Each monomer is 223 amino acids and symmetrically bind both sides of the unique palindromic nucleotide sequence AGATCT, cleaving the scissile phosphodiester bond between the first Adenine and Guanine nucleotides on both strands of the DNA molecule, creating sticky ends with 5' end overhangs.
Being a type II restriction enzyme, BglII does not require ATP (adenosine triphosphate) for its enzymatic function, but only requires association with a divalent metal cation, most likely Mg2+. Unlike other restriction enzymes of its class, BglII has been shown to possess some unique structural characteristics, such as a β-sandwich subdomain, and appears to undergo a unique conformational change upon dimerization,[2] but its overall structure and mechanism of catalysis remain consistent with other type II restriction enzymes.
Restriction endonucleases play a very important role in modern molecular cloning techniques. Because of their unique recognition/cut sites, restriction enzymes can be used to precisely cut DNA at specific locations in a predictable manner. Once cut, the DNA (usually) possesses so-called "sticky ends", which can then allow the DNA fragment to hybridise into a DNA vector. Ligating enzymes are used to covalently link the desired fragment to the vector for subsequent DNA cloning.
| Identifiers | |
| Name | BglII Restriction Endonuclease |
| Entrez | 6173168 |
| PDB | 1DFM |
| ACCESSION # | Q45488 |
| EC Number | 3.1.21.4 |
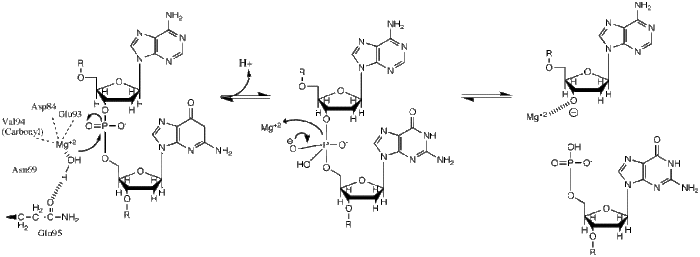
Mechanism
BglII catalyses phosphodiester bond cleavage at the DNA backbone through a phosphoryl transfer to water.[1] Studies on the mechanism of restriction enzymes have revealed several general features that seem to be true in almost all cases, although the actual mechanism for each enzyme is most likely some variation of this general mechanism. This mechanism requires a base to generate the hydroxide ion from water, which will act as the nucleophile and attack the phosphorus in the phosphodiester bond. Also required is a Lewis acid to stabilize the extra negative charge of the pentacoordinated transition state phosphorus, as well as a general acid or metal ion that stabilizes the leaving group (3’-O−).
Structure

Although restriction endonucleases show little sequence similarity, crystal structures reveal that they all share a highly similar α/β core consisting of a six-stranded β-sheet flanked by five α-helices, two of which mediate dimerization.[1] This core carries the active site (catalytic center) and the residues that contact DNA in the major groove. BglII is unique in that its α/β core is augmented by a β-sandwich subdomain that has several projections that extend outward to grip the DNA, allowing BglII to completely encircle the DNA molecule. This atypical feature of BglII suggests a unique hinge motion for DNA binding and release.[2] Comparative structural studies of the free enzyme vs. the BglII-DNA complex showed that the enzyme opens by a dramatic scissor-like motion, accompanied by a complete rearrangement of the α-helices at the dimer interface. These structural studies also revealed that within each monomer a set of residues lowers or raises to alternatively sequester or expose the active site residues. These dramatic differences in structure in the free vs. bound enzyme have yet to be observed in any other restriction endonuclease and may possibly represent a novel mechanism for capturing DNA that may extend to other proteins that encircle DNA.[2][3]
Active site

Structural studies of endonucleases have revealed a similar architecture for the active site with the residues following the weak consensus sequence Glu/Asp-(X)9-20-Glu/Asp/Ser-X-Lys/Glu. BglII's active site is similar to other endonucleases', following the sequence Asp-(X)9-Glu-X-Gln. In its active site there sits a divalent metal cation, most likely Mg2+, that interacts with Asp-84, Val-94, a phosphoryl oxygen, and three water molecules. One of these water molecules, is able act as a nucleophile because of its proximity to the scissile phosphoryl (its orientation being fixed by a hydrogen bond with the side chain amide oxygen of Gln-95[1][4]) and its contact with the metal cation (which lowers its pKa, promoting the water's nucleophilicity).
See also
- BamHI, a nuclease enzyme from Bacillus amyloliquefaciens
- FokI, a nuclease enzyme from Flavobacterium okeanokoites
- EcoRI, a nuclease enzyme from E. coli
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution". Nature Structural Biology 7 (2): 134–40. February 2000. doi:10.1038/72405. PMID 10655616.
- ↑ 2.0 2.1 2.2 "Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease". Nature Structural Biology 8 (2): 126–30. February 2001. doi:10.1038/84111. PMID 11175900.
- ↑ "Restriction endonucleases: one of these things is not like the others". Nature Structural Biology 7 (2): 89–91. February 2000. doi:10.1038/72450. PMID 10655603.
- ↑ 4.0 4.1 "Structure and function of type II restriction endonucleases". Nucleic Acids Research 29 (18): 3705–27. September 2001. doi:10.1093/nar/29.18.3705. PMID 11557805.
External links
- Restriction Enzyme Database
- NCBI Protein Database Entry
- Structure Summary, MMDB
- BglII, Biology.Kenyon.edu
 |