Biology:Bioluminescent bacteria

Bioluminescent bacteria are light-producing bacteria that are predominantly present in sea water, marine sediments, the surface of decomposing fish and in the gut of marine animals. While not as common, bacterial bioluminescence is also found in terrestrial and freshwater bacteria.[1] These bacteria[clarification needed] may be free living (such as Vibrio harveyi) or in symbiosis with animals such as the Hawaiian Bobtail squid (Aliivibrio fischeri) or terrestrial nematodes (Photorhabdus luminescens). The host organisms provide these bacteria[clarification needed] a safe home and sufficient nutrition. In exchange, the hosts use the light produced by the bacteria for camouflage, prey and/or mate attraction. Bioluminescent bacteria have evolved symbiotic relationships with other organisms in which both participants benefit close to equally.[2] Another possible reason bacteria use luminescence reaction is for quorum sensing, an ability to regulate gene expression in response to bacterial cell density.[3]
History
Records of bioluminescence due to bacteria have existed for thousands of years.[4] They appear in the folklore of many regions, including Scandinavia and the Indian subcontinent. Both Aristotle and Charles Darwin have described the phenomenon of the oceans glowing.[4] Since its discovery less than 30 years ago, the enzyme luciferase and its regulatory gene, lux, have led to major advances in molecular biology, through use as a reporter gene.[5] Luciferase was first purified by McElroy and Green in 1955.[6] It was later discovered that there were two subunits to luciferase, called subunits α and β. The genes encoding these enzymes, luxA and luxB, respectively, were first isolated in the lux operon of Aliivibrio fisheri[4].
Purpose of bio-luminescence
The wide-ranged biological purposes of bio-luminescence include but are not limited to attraction of mates,[7] defense against predators, and warning signals. In the case of bioluminescent bacteria, bio-luminescence mainly serves as a form of dispersal. It has been hypothesized that enteric bacteria (bacteria that survive in the guts of other organisms) - especially those prevalent in the depths of the ocean - employ bio-luminescence as an effective form of distribution.[8] After making their way into the digestive tracts of fish and other marine organisms and being excreted in fecal pellets, bioluminescent bacteria are able to utilize their bio-luminescent capabilities to lure in other organisms and prompt ingestion of these bacterial-containing fecal pellets.[9] The bio-luminescence of bacteria thereby ensures their survival, persistence, and dispersal as they are able to enter and inhabit other organisms.
Regulation of bio-luminescence
The regulation of bio-luminescence in bacteria is achieved through the regulation of the oxidative enzyme called luciferase. It is important that bio-luminescent bacteria decrease production rates of luciferase when the population is sparse in number in order to conserve energy. Thus, bacterial bioluminescence is regulated by means of chemical communication referred to as quorum sensing.[10] Essentially, certain signaling molecules named autoinducers[11] with specific bacterial receptors become activated when the population density of bacteria is high enough. The activation of these receptors leads to a coordinated induction of luciferase production that ultimately yields visible luminescence.[12]
Biochemistry of bio-luminescence

The chemical reaction that is responsible for bio-luminescence is catalyzed by the enzyme luciferase. In the presence of oxygen, luciferase catalyzes the oxidation of an organic molecule called luciferin.[13] Though bio-luminescence across a diverse range of organisms such as bacteria, insects, and dinoflagellates function in this general manner (utilizing luciferase and luciferin), there are different types of luciferin-luciferase systems. For bacterial bio-luminescence specifically, the biochemical reaction involves the oxidation of an aliphatic aldehyde by a reduced flavin mononucleotide.[14] The products of this oxidation reaction include an oxidized flavin mononucleotide, a fatty acid chain, and energy in the form of a blue-green visible light.[15]
Reaction: FMNH2 + O2 + RCHO → FMN + RCOOH + H2O + light
Evolution of bio-luminescence
Of all light emitters in the ocean, bio-luminescent bacteria is the most abundant and diverse. However, the distribution of bio-luminescent bacteria is uneven, which suggests evolutionary adaptations. The bacterial species in terrestrial genera such as Photorhabdus are bio-luminescent. On the other hand, marine genera with bio-luminescent species such as Vibrio and Shewanella oneidensis have different closely related species that are not light emitters.[16] Nevertheless, all bio-luminescent bacteria share a common gene sequence: the enzymatic oxidation of Aldehyde and reduced Flavin mononucleotide by luciferase which are contained in the lux operon.[17] Bacteria from distinct ecological niches contain this gene sequence; therefore, the identical gene sequence evidently suggests that bio-luminescence bacteria result from evolutionary adaptations.
Use as laboratory tool
After the discovery of the lux operon, the use of bioluminescent bacteria as a laboratory tool is claimed to have revolutionized the area of environmental microbiology.[4] The applications of bioluminescent bacteria include biosensors for detection of contaminants, measurement of pollutant toxicity [4][18][19] and monitoring of genetically engineered bacteria released into the environment.[20][21][22] Biosensors, created by placing a lux gene construct under the control of an inducible promoter, can be used to determine the concentration of specific pollutants.[4] Biosensors are also able to distinguish between pollutants that are bioavailable and those that are inert and unavailable.[4] For example, Pseudomonas fluorescens has been genetically engineered to be capable of degrading salicylate and naphthalene, and is used as a biosensor to assess the bioavailability of salicylate and naphthalene.[4] Biosensors can also be used as an indicator of cellular metabolic activity and to detect the presence of pathogens.[4]
Evolution
The light-producing chemistry behind bioluminescence varies across the lineages of bioluminescent organisms.[16] Based on this observation, bioluminescence is believed to have evolved independently at least 40 times.[16] In bioluminescent bacteria, the reclassification of the members ofVibrio fischeri species group as a new genus, Aliivibrio, has led to increased interest in the evolutionary origins of bioluminescence[16]. Among bacteria, the distribution of bioluminescent species is polyphyletic. For instance, while all species in the terrestrial genus Photorhabdus are luminescent, the genera Aliivibrio, Photobacterium, Shewanella and Vibrio contain both luminous and non-luminous species.[16] Despite bioluminescence in bacteria not sharing a common origin, they all share a gene sequence in common. The appearance of the highly conserved lux operon in bacteria from very different ecological niches suggests a strong selective advantage despite the high energetic costs of producing light. DNA repair is thought to be the initial selective advantage for light production in bacteria.[16] Consequently, the lux operon may have been lost in bacteria that evolved more efficient DNA repair systems but retained in those where visible light became a selective advantage.[16][23] The evolution of quorum sensing is believed to have afforded further selective advantage for light production. Quorum sensing allows bacteria to conserve energy by ensuring that they do not synthesize light-producing chemicals unless a sufficient concentration are present to be visible.[16]
Bacterial groups that exhibit bioluminescence
All bacterial species that have been reported to possess bioluminescence belong within the families Vibrionaceae, Shewanellaceae, or Enterobacteriaceae, all of which are assigned to the class Gammaproteobacteria.[24]
| Family | Genus | Species |
|---|---|---|
| Enterobacteriaceae | Photorhabdus | Photorhabdus asymbiotica |
| Shewanellaceae | Shewanella | Shewanella woodyi
Shewanella hanedai |
| Vibrionaceae | Aliivibrio | Aliivibrio fischeri
Aliivibrio logei Aliivibrio salmonicida Aliivibrio sifiae Aliivibrio "thorii" Aliivibrio wodanis |
| Photobacterium | Photobacterium aquimaris
Photobacterium kishitanii Photobacterium mandapamensis | |
| Vibrio | Vibrio azureus
Vibrio "beijerinckii" Vibrio chagasii Vibrio mediterranei Vibrio orientalis Vibrio sagamiensis Vibrio splendidus | |
| "Candidatus Photodesmus" | "Candidatus Photodesmus katoptron" |
(List from Dunlap and Henryk (2013), "Luminous Bacteria", The Prokaryotes [24])
Distribution
Bioluminescent bacteria are most abundant in marine environments during spring blooms when there are high nutrient concentrations. These light-emitting organisms are found mainly in coastal waters near the outflow of rivers, such as the northern Adriatic Sea, Gulf of Trieste, northwestern part of the Caspian Sea, coast of Africa and many more.[25] These are known as milky seas. Bioluminescent bacteria are also found in freshwater and terrestrial environments but are less wide spread than in seawater environments. They are found globally, as free-living, symbiotic or parasitic forms [1] and possibly as opportunistic pathogens.[24] Factors that affect the distribution of bioluminescent bacteria include temperature, salinity, nutrient concentration, pH level and solar radiation.[26] For example, Aliivibrio fischeri grows favourably in environments that have temperatures between 5 and 30 °C and a pH that is less than 6.8; whereas, Photobacterium phosphoreum thrives in conditions that have temperatures between 5 and 25 °C and a pH that is less than 7.0.[27]
Genetic diversity
All bioluminescent bacteria share a common gene sequence: the lux operon characterized by the luxCDABE gene organization.[24] LuxAB codes for luciferase while luxCDE codes for a fatty-acid reductase complex that is responsible for synthesizing aldehydes for the bioluminescent reaction. Despite this common gene organization, variations, such as the presence of other lux genes, can be observed among species. Based on similarities in gene content and organization, the lux operon can be organized into the following four distinct types: the Aliivibrio/Shewanella type, the Photobacterium type, theVibrio/Candidatus Photodesmus type, and the Photorhabdus type. While this organization follows the genera classification level for members of Vibrionaceae (Aliivibrio, Photobacterium, and Vibrio), its evolutionary history is not known.[24]
With the exception of the Photorhabdus operon type, all variants of the lux operon contain the flavin reductase-encoding luxG gene.[24] Most of the Aliivibrio/Shewanella type operons contain additional luxI/luxR regulatory genes that are used for autoinduction during quorum sensing.[28] The Photobacterum operon type is characterized by the presence of rib genes that code for riboflavin, and forms the lux-rib operon. TheVibrio/Candidatus Photodesmus operon type differs from both the Aliivibrio/Shewanella and the Photobacterium operon types in that the operon has no regulatory genes directly associated with it.[24]
Mechanism
All bacterial luciferases are approximately 80 KDa heterodimers containing two subunits: α and β. The α subunit is responsible for light emission.[4] The luxA and luxB genes encode for the α and β subunits, respectively. In most bioluminescent bacteria, the luxA and luxB genes are flanked upstream by luxC and luxD and downstream by luxE.[4]
The bioluminescent reaction is as follows:
FMNH2 + O2 + R-CHO -> FMN + H2O + R-COOH + Light (~ 495 nm)
Molecular oxygen reacts with FMNH2 (reduced flavin mononucleotide) and a long-chain aldehyde to produce FMN (flavin mononucleotide), water and a corresponding fatty acid. The blue-green light emission of bioluminescence, such as that produced by Photobacterium phosphoreum and Vibro harveyi, results from this reaction.[4] Because light emission involves expending six ATP molecules for each photon, it is an energetically expensive process. For this reason, light emission is not constitutively expressed in bioluminescent bacteria; it is expressed only when physiologically necessary.
Quorum sensing

Bioluminescence in bacteria can be regulated through a phenomenon known as autoinduction or quorum sensing.[4] Quorum sensing is a form of cell-to-cell communication that alters gene expression in response to cell density. Autoinducer is a diffusible pheromone produced constitutively by bioluminescent bacteria and serves as an extracellular signalling molecule.[4] When the concentration of autoinducer secreted by bioluminescent cells in the environment reaches a threshold (above 107 cells per mL), it induces the expression of luciferase and other enzymes involved in bioluminescence.[4] Bacteria are able to estimate their density by sensing the level of autoinducer in the environment and regulate their bioluminescence such that it is expressed only when there is a sufficiently high cell population. A sufficiently high cell population ensures that the bioluminescence produced by the cells will be visible in the environment.
A well known example of quorum sensing is that which occurs between Aliivibrio fischeri and its host. This process is regulated by LuxI and LuxR, encoded by luxI and luxR respectively. LuxI is autoinducer synthase that produces autoinducer (AI) while LuxR functions as both a receptor and transcription factor for the lux operon.[4] When LuxR binds AI, LuxR-AI complex activates transcription of the lux operon and induces the expression of luciferase.[28] Using this system, A. fischeri has shown that bioluminescence is expressed only when the bacteria are host-associated and have reached sufficient cell densities.[29]
Another example of quorum sensing by bioluminescent bacteria is by Vibrio harveyi, which are known to be free-living. Unlike Aliivibrio fischeri, V. harveyi do not possess the luxI/luxR regulatory genes and therefore have a different mechanism of quorum sensing regulation. Instead, they use the system known as three-channel quorum sensing system.[30] Vibrio use small non-coding RNAs called Qrr RNAs to regulate quorum sensing, using them to control translation of energy-costly molecules.
Role
The uses of bioluminescence and its biological and ecological significance for animals, including host organisms for bacteria symbiosis, have been widely studied. The biological role and evolutionary history for specifically bioluminescent bacteria still remains quite mysterious and unclear.[4][31] However, there are continually new studies being done to determine the impacts that bacterial bioluminescence can have on our constantly changing environment and society. Aside from the many scientific and medical uses, scientists have also recently begun to come together with artists and designers to explore new ways of incorporating bioluminescent bacteria, as well as bioluminescent plants, into urban light sources to reduce the need for electricity.[32] They have also begun to use bioluminescent bacteria as a form of art and urban design for the wonder and enjoyment of human society.[33][34][35]
One explanation for the role of bacterial bioluminescence is from the biochemical aspect. Several studies have shown the biochemical roles of the luminescence pathway. It can function as an alternate pathway for electron flow under low oxygen concentration, which can be advantageous when no fermentable substrate is available.[1] In this process, light emission is a side product of the metabolism.
Evidence also suggests that bacterial luciferase contributes to the resistance of oxidative stress. In laboratory culture, luxA and luxB mutants of Vibrio harveyi, which lacked luciferase activity, showed impairment of growth under high oxidative stress compared to wild type. The luxD mutants, which had an unaffected luciferase but were unable to produce luminescence, showed little or no difference. This suggests that luciferase mediates the detoxification of reactive oxygen.[36]
Bacterial bioluminescence has also been proposed to be a source of internal light in photoreactivation, a DNA repair process carried out by photolyase.[37] Experiments have shown that non-luminescent V. harveyi mutants are more sensitive to UV irradiation, suggesting the existence of a bioluminescent-mediated DNA repair system.[23]
Another hypothesis, called the “bait hypothesis”, is that bacterial bioluminescence attracts predators who will assist in their dispersal.[37] They are either directly ingested by fish or indirectly ingested by zooplankton that will eventually be consumed by higher trophic levels. Ultimately, this may allow passage into the fish gut, a nutrient-rich environment where the bacteria can divide, be excreted, and continue their cycle. Experiments using luminescent Photobacterium leiognathi and non-luminescent mutants have shown that luminescence attracts zooplankton and fish, thus supporting this hypothesis.[37]
Symbiosis with other organisms
The symbiotic relationship between the Hawaiian bobtail squid Euprymna scolopes and the marine gram-negative bacterium Aliivibrio fischeri has been well studied. The two organisms exhibit a mutualistic relationship in which bioluminescence produced by A. fischeri helps to attract pray to the squid host, which provides nutrient-rich tissues and a protected environment forA. fischeri.[38] Bioluminescence provided by A. fischeri also aids in the defense of the squid E. scolopes by providing camouflage during its nighttime foraging activity.[39] Following bacterial colonization, the specialized organs of the squid undergo developmental changes and a relationship becomes established. The squid expels 90% of the bacterial population each morning, because it no longer needs to produce bioluminescence in the daylight.[4] This expulsion benefits the bacteria by aiding in their dissemination. A single expulsion by one bobtail squid produces enough bacterial symbionts to fill 10,000m3 of seawater at a concentration that is comparable to what is found in coastal waters.[39] Thus, in at least some habitats, the symbiotic relationship between A. fischeri and E. scolopes plays a key role in determining the abundance and distribution of E. scolopes. There is a higher abundance of A. fischeri in the vicinity of a population of E. scolopes and this abundance markedly decreases with increasing distance from the host's habitat.[39]
Bioluminescent Photobacterium species also engage in mutually beneficial associations with fish and squid.[40] Dense populations of P. kishitanii, P. leiogathi, and P. mandapamensis can live in the light organs of marine fish and squid, and are provided with nutrients and oxygen for reproduction[40] in return for providing bioluminescence to their hosts, which can aid in sex-specific signaling, predator avoidance, locating or attracting prey, and schooling.[40]
| In the table below, the images at the right indicate in blue the locations of the light organ of different families of symbiotically luminous fish and squid.[41] E indicates an external expulsion of the bioluminescent bacteria directly into the seawater. I indicates an internal expulsion of the bioluminescent bacteria in the digestive tract. (E) or (I) indicate a putative localization of the expulsion.[42] |
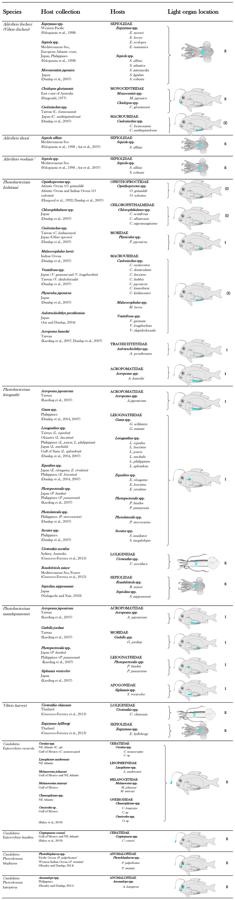
|
See also
- Bioluminescence
- Bioluminescent shunt hypothesis
- List of bioluminescent organisms
References
- ↑ 1.0 1.1 1.2 Nealson, K. H.; Hastings, J. W. (1979-12-01). "Bacterial bioluminescence: its control and ecological significance". Microbiological Reviews 43 (4): 496–518. doi:10.1128/mmbr.43.4.496-518.1979. ISSN 0146-0749. PMID 396467.
- ↑ McFall-Ngai, Margaret; Heath-Heckman, Elizabeth A.C.; Gillette, Amani A.; Peyer, Suzanne M.; Harvie, Elizabeth A. (2012). "The secret languages of coevolved symbioses: Insights from the Euprymna scolopes–Vibrio fischeri symbiosis". Seminars in Immunology 24 (1): 3–8. doi:10.1016/j.smim.2011.11.006. PMID 22154556.
- ↑ Waters, Christopher M.; Bassler, Bonnie L. (2005-10-07). "QUORUM SENSING: Cell-to-Cell Communication in Bacteria" (in en). Annual Review of Cell and Developmental Biology 21 (1): 319–346. doi:10.1146/annurev.cellbio.21.012704.131001. PMID 16212498.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 Nunes-Halldorson, Vânia da Silva; Duran, Norma Letícia (2003-06-01). "Bioluminescent bacteria: lux genes as environmental biosensors". Brazilian Journal of Microbiology 34 (2): 91–96. doi:10.1590/S1517-83822003000200001. ISSN 1517-8382.
- ↑ "Luciferase Reporters" (in en). https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/luciferase-reporters.html.
- ↑ McElroy, W.D.; Green (1954). "Enzymatic properties of bacterial luciferase". Archives of Biochemistry and Biophysics 56 (1): 240–255. doi:10.1016/0003-9861(55)90353-2. PMID 14377570.
- ↑ Herring, Peter J. (2007-08-01). "REVIEW. Sex with the lights on? A review of bioluminescent sexual dimorphism in the sea". Journal of the Marine Biological Association of the United Kingdom 87 (4): 829–842. doi:10.1017/S0025315407056433. ISSN 1469-7769. https://www.cambridge.org/core/journals/journal-of-the-marine-biological-association-of-the-united-kingdom/article/div-classtitlereview-sex-with-the-lights-on-a-review-of-bioluminescent-sexual-dimorphism-in-the-seadiv/5F913C5E66B6CC36E0CDE66D4D555DA9.
- ↑ Ruby, E. G.; Morin, J. G. (1979-09-01). "Luminous enteric bacteria of marine fishes: a study of their distribution, densities, and dispersion". Applied and Environmental Microbiology 38 (3): 406–411. doi:10.1128/AEM.38.3.406-411.1979. ISSN 0099-2240. PMID 16345429. Bibcode: 1979ApEnM..38..406R.
- ↑ Ruby, E. G.; Morin, J. G. (1979-09-01). "Luminous Enteric Bacteria of Marine Fishes: a Study of Their Distribution, Densities, and Dispersion" (in en). Applied and Environmental Microbiology 38 (3): 406–411. doi:10.1128/AEM.38.3.406-411.1979. ISSN 0099-2240. PMID 16345429. Bibcode: 1979ApEnM..38..406R.
- ↑ Wilson, Thérèse; Hastings, J. Woodland (2003-11-28). "BIOLUMINESCENCE" (in en). Annual Review of Cell and Developmental Biology 14 (1): 197–230. doi:10.1146/annurev.cellbio.14.1.197. PMID 9891783.
- ↑ Eberhard, A.; Burlingame, A. L.; Eberhard, C.; Kenyon, G. L.; Nealson, K. H.; Oppenheimer, N. J. (1981-04-01). "Structural identification of autoinducer of Photobacterium fischeri luciferase". Biochemistry 20 (9): 2444–2449. doi:10.1021/bi00512a013. ISSN 0006-2960. PMID 7236614.
- ↑ Pérez-Velázquez, Judith; Gölgeli, Meltem; García-Contreras, Rodolfo (2016-08-25). "Mathematical Modelling of Bacterial Quorum Sensing: A Review" (in en). Bulletin of Mathematical Biology 78 (8): 1585–1639. doi:10.1007/s11538-016-0160-6. ISSN 0092-8240. PMID 27561265. http://repository.bilkent.edu.tr/bitstream/11693/36891/1/bilkent-research-paper.pdf.
- ↑ Kaskova, Zinaida M.; Tsarkova, Aleksandra S.; Yampolsky, Ilia V. (2016-10-24). "1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine" (in en). Chem. Soc. Rev. 45 (21): 6048–6077. doi:10.1039/c6cs00296j. ISSN 1460-4744. PMID 27711774.
- ↑ Wiles, Siouxsie; Ferguson, Kathryn; Stefanidou, Martha; Young, Douglas B.; Robertson, Brian D. (2005-07-01). "Alternative Luciferase for Monitoring Bacterial Cells under Adverse Conditions" (in en). Applied and Environmental Microbiology 71 (7): 3427–3432. doi:10.1128/AEM.71.7.3427-3432.2005. ISSN 0099-2240. PMID 16000745. Bibcode: 2005ApEnM..71.3427W.
- ↑ Fisher, Andrew J.; Thompson, Thomas B.; Thoden, James B.; Baldwin, Thomas O.; Rayment, Ivan (1996-09-06). "The 1.5-Å Resolution Crystal Structure of Bacterial Luciferase in Low Salt Conditions" (in en). Journal of Biological Chemistry 271 (36): 21956–21968. doi:10.1074/jbc.271.36.21956. ISSN 0021-9258. PMID 8703001.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 Widder, E. A. (2010-05-07). "Bioluminescence in the Ocean: Origins of Biological, Chemical, and Ecological Diversity" (in en). Science 328 (5979): 704–708. doi:10.1126/science.1174269. ISSN 0036-8075. PMID 20448176. Bibcode: 2010Sci...328..704W.
- ↑ O'Kane, Dennis J.; Prasher, Douglas C. (1992-02-01). "Evolutionary origins of bacterial bioluminescence" (in en). Molecular Microbiology 6 (4): 443–449. doi:10.1111/j.1365-2958.1992.tb01488.x. ISSN 1365-2958. PMID 1560772.
- ↑ Ptitsyn, L. R.; Horneck, G.; Komova, O.; Kozubek, S.; Krasavin, E. A.; Bonev, M.; Rettberg, P. (1997-11-01). "A biosensor for environmental genotoxin screening based on an SOS lux assay in recombinant Escherichia coli cells". Applied and Environmental Microbiology 63 (11): 4377–4384. doi:10.1128/AEM.63.11.4377-4384.1997. ISSN 0099-2240. PMID 9361425. Bibcode: 1997ApEnM..63.4377P.
- ↑ Widder, Edith A.; Falls, Beth (March 2014). "Review of Bioluminescence for Engineers and Scientists in Biophotonics". IEEE Journal of Selected Topics in Quantum Electronics 20 (2): 232–241. doi:10.1109/JSTQE.2013.2284434. ISSN 1077-260X. Bibcode: 2014IJSTQ..20..232W.
- ↑ Mahaffee, W. F.; Bauske, E. M.; van Vuurde, J. W.; van der Wolf, J. M.; van den Brink, M.; Kloepper, J. W. (1997-04-01). "Comparative analysis of antibiotic resistance, immunofluorescent colony staining, and a transgenic marker (bioluminescence) for monitoring the environmental fate of rhizobacterium". Applied and Environmental Microbiology 63 (4): 1617–1622. doi:10.1128/AEM.63.4.1617-1622.1997. ISSN 0099-2240. PMID 9097457. Bibcode: 1997ApEnM..63.1617M.
- ↑ Shaw, J. J.; Dane, F.; Geiger, D.; Kloepper, J. W. (1992-01-01). "Use of bioluminescence for detection of genetically engineered microorganisms released into the environment". Applied and Environmental Microbiology 58 (1): 267–273. doi:10.1128/AEM.58.1.267-273.1992. ISSN 0099-2240. PMID 1311542. Bibcode: 1992ApEnM..58..267S.
- ↑ Flemming, C. A.; Lee, H.; Trevors, J. T. (1994-09-01). "Bioluminescent Most-Probable-Number Method To Enumerate lux-Marked Pseudomonas aeruginosa UG2Lr in Soil". Applied and Environmental Microbiology 60 (9): 3458–3461. doi:10.1128/AEM.60.9.3458-3461.1994. ISSN 0099-2240. PMID 16349396. Bibcode: 1994ApEnM..60.3458F.
- ↑ 23.0 23.1 Czyz, A.; Wróbel, B.; Wegrzyn, G. (2000-02-01). "Vibrio harveyi bioluminescence plays a role in stimulation of DNA repair". Microbiology 146 (2): 283–288. doi:10.1099/00221287-146-2-283. ISSN 1350-0872. PMID 10708366.
- ↑ 24.0 24.1 24.2 24.3 24.4 24.5 24.6 Delong, Edward F.; Stackebrandt, Erko; Lory, Stephen; Thompson, Fabiano (2013) (in en). The Prokaryotes - Springer. doi:10.1007/978-3-642-31331-8. ISBN 978-3-642-31330-1.
- ↑ Staples, Robert F.; States., United (1966) (in en). Details - The distribution and characteristics of surface bioluminescence in the oceans / Robert F. Staples. - Biodiversity Heritage Library. doi:10.5962/bhl.title.46935. https://www.biodiversitylibrary.org/bibliography/46935.
- ↑ Soto, W.; Gutierrez, J.; Remmenga, M. D.; Nishiguchi, M. K. (2009-01-01). "Salinity and Temperature Effects on Physiological Responses of Vibrio fischeri from Diverse Ecological Niches" (in en). Microbial Ecology 57 (1): 140–150. doi:10.1007/s00248-008-9412-9. ISSN 0095-3628. PMID 18587609.
- ↑ Waters, Paul; Lloyd, David (1985). "Salt, pH and temperature dependencies of growth and bioluminescence of three species of luminous bacteria analysed on gradient plates". Journal of General Microbiology 131 (11): 65–9. doi:10.1099/00221287-131-11-2865.
- ↑ 28.0 28.1 Bassler, B. L. (1999-12-01). "How bacteria talk to each other: regulation of gene expression by quorum sensing". Current Opinion in Microbiology 2 (6): 582–587. doi:10.1016/s1369-5274(99)00025-9. ISSN 1369-5274. PMID 10607620.
- ↑ Fuqua, W. C.; Winans, S. C.; Greenberg, E. P. (1994-01-01). "Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators". Journal of Bacteriology 176 (2): 269–275. doi:10.1128/jb.176.2.269-275.1994. ISSN 0021-9193. PMID 8288518.
- ↑ Defoirdt, Tom; Boon, Nico; Sorgeloos, Patrick; Verstraete, Willy; Bossier, Peter (2008-01-01). "Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work". The ISME Journal 2 (1): 19–26. doi:10.1038/ismej.2007.92. ISSN 1751-7362. PMID 18180744.
- ↑ Vannier, Thomas; Hingamp, Pascal; Turrel, Floriane; Tanet, Lisa; Lescot, Magali; Timsit, Youri (2020-06-01). "Diversity and evolution of bacterial bioluminescence genes in the global ocean" (in en). NAR Genomics and Bioinformatics 2 (2): lqaa018. doi:10.1093/nargab/lqaa018. PMID 33575578. PMC 7671414. https://academic.oup.com/nargab/article/2/2/lqaa018/5805306.
- ↑ Ardavani, Olympia; Zerefos, Stelios; Doulos, Lambros T. (2020-01-01). "Redesigning the exterior lighting as part of the urban landscape: The role of transgenic bioluminescent plants in mediterranean urban and suburban lighting environments" (in en). Journal of Cleaner Production 242: 118477. doi:10.1016/j.jclepro.2019.118477. ISSN 0959-6526.
- ↑ Latz, Michael I. (2019). "The Making of Infinity Cube, a Bioluminescence Art Exhibit" (in en). Limnology and Oceanography Bulletin 28 (4): 130–134. doi:10.1002/lob.10345. ISSN 1539-6088.
- ↑ DYBAS, CHERYL LYN (2019). "A New Wave SCIENCE AND ART MEET IN THE SEA". Oceanography 32 (1): 10–12. ISSN 1042-8275.
- ↑ "More-than-Human Media Architecture" (in EN). Proceedings of the 4th Media Architecture Biennale Conference. doi:10.1145/3284389.3284495. https://eprints.qut.edu.au/121705/41/121705.pdf.
- ↑ Szpilewska, Hanna; Czyz, Agata; Wegrzyn, Grzegorz (2003-11-01). "Experimental evidence for the physiological role of bacterial luciferase in the protection of cells against oxidative stress". Current Microbiology 47 (5): 379–382. doi:10.1007/s00284-002-4024-y. ISSN 0343-8651. PMID 14669913.
- ↑ 37.0 37.1 37.2 Zarubin, Margarita; Belkin, Shimshon; Ionescu, Michael; Genin, Amatzia (2012-01-17). "Bacterial bioluminescence as a lure for marine zooplankton and fish". Proceedings of the National Academy of Sciences of the United States of America 109 (3): 853–857. doi:10.1073/pnas.1116683109. ISSN 1091-6490. PMID 22203999. Bibcode: 2012PNAS..109..853Z.
- ↑ Naughton, Lynn M.; Mandel, Mark J. (2012-03-01). "Colonization of Euprymna scolopes Squid by Vibrio fischeri". Journal of Visualized Experiments (61): e3758. doi:10.3791/3758. ISSN 1940-087X. PMID 22414870.
- ↑ 39.0 39.1 39.2 Ruby, E. G.; Lee, K. H. (1998-03-01). "The Vibrio fischeri-Euprymna scolopes Light Organ Association: Current Ecological Paradigms". Applied and Environmental Microbiology 64 (3): 805–812. doi:10.1128/AEM.64.3.805-812.1998. ISSN 0099-2240. PMID 16349524. Bibcode: 1998ApEnM..64..805R.
- ↑ 40.0 40.1 40.2 Urbanczyk, Henryk; Ast, Jennifer C.; Dunlap, Paul V. (2011-03-01). "Phylogeny, genomics, and symbiosis of Photobacterium" (in en). FEMS Microbiology Reviews 35 (2): 324–342. doi:10.1111/j.1574-6976.2010.00250.x. ISSN 0168-6445. PMID 20883503.
- ↑ Nealson, K. H. and Hastings, J. W. (1979) "Bacterial bioluminescence: its control and ecological significance", Microbiol. Rev., 43: 496–518. doi:10.1128/MMBR.43.4.496-518.1979.
- ↑ Tanet, Lisa; Martini, Séverine; Casalot, Laurie; Tamburini, Christian (2020). "Reviews and syntheses: Bacterial bioluminescence – ecology and impact in the biological carbon pump". Biogeosciences 17 (14): 3757–3778. doi:10.5194/bg-17-3757-2020. Bibcode: 2020BGeo...17.3757T.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Further reading
- Hastings, J W; Nealson, K H (1977). "Bacterial Bioluminescence". Annual Review of Microbiology 31 (1): 549–595. doi:10.1146/annurev.mi.31.100177.003001. ISSN 0066-4227. PMID 199107.
 |

