Biology:Bromodomain
| Bromodomain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
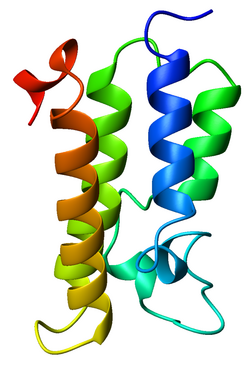 Ribbon diagram of the GCN5 bromodomain from Saccharomyces cerevisiae, colored from blue (N-terminus) to red (C-terminus).[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | Bromodomain | ||||||||||
| Pfam | PF00439 | ||||||||||
| InterPro | IPR001487 | ||||||||||
| SMART | SM00297 | ||||||||||
| PROSITE | PDOC00550 | ||||||||||
| SCOP2 | 1b91 / SCOPe / SUPFAM | ||||||||||
| CDD | cd04369 | ||||||||||
| |||||||||||
A bromodomain is an approximately 110 amino acid protein domain that recognizes acetylated lysine residues, such as those on the N-terminal tails of histones. Bromodomains, as the "readers" of lysine acetylation, are responsible in transducing the signal carried by acetylated lysine residues and translating it into various normal or abnormal phenotypes.[2] Their affinity is higher for regions where multiple acetylation sites exist in proximity. This recognition is often a prerequisite for protein-histone association and chromatin remodeling. The domain itself adopts an all-α protein fold, a bundle of four alpha helices each separated by loop regions of variable lengths that form a hydrophobic pocket that recognizes the acetyl lysine.[1][3]
Discovery
The bromodomain was identified as a novel structural motif by John W. Tamkun and colleagues studying the Drosophila gene Brahma/brm, and showed sequence similarity to genes involved in transcriptional activation.[4] The name "bromodomain" is derived from the relationship of this domain with Brahma and is unrelated to the chemical element bromine.
Bromodomain-containing proteins
Bromodomain-containing proteins can have a wide variety of functions, ranging from histone acetyltransferase activity and chromatin remodeling to transcriptional mediation and co-activation. Of the 43 known in 2015, 11 had two bromodomains, and one protein had 6 bromodomains.[2] Preparation, biochemical analysis, and structure determination of the bromodomain containing proteins have been described in detail.[5]
Bromo- and Extra-Terminal domain (BET) family
A well-known example of a bromodomain family is the BET (Bromodomain and extraterminal domain) family. Members of this family include BRD2, BRD3, BRD4 and BRDT.[6]
Other
However proteins such as ASH1L also contain a bromodomain. Dysfunction of BRD proteins has been linked to diseases such as human squamous cell carcinoma and other forms of cancer.[7] Histone acetyltransferases, including EP300 and PCAF, have bromodomains in addition to acetyl-transferase domains.[8][9][10]
Not considered part of the BET family (yet containing a bromodomain) are BRD7, and BRD9.
Role in human disease
The role of bromodomains in translating a deregulated cell acetylome into disease phenotypes was recently unveiled by the development of small molecule bromodomain inhibitors. This breakthrough discovery highlighted bromodomain-containing proteins as key players in cancer biology, as well as inflammation[11] and remyelination in multiple sclerosis.[2]
Members of the BET family have been implicated as targets in both human cancer[12][13] and multiple sclerosis.[14] BET inhibitors have shown therapeutic effects in multiple preclinical models of cancer and are currently in clinical trials in the United States.[15] Their application in multiple sclerosis is still in the preclinical stage.
Small molecule inhibitors of non-BET bromodomain proteins BRD7 and BRD9 have also been developed.[16][17]
See also
- Chromodomain
- BET inhibitor
- BRD2
- BRD3
- BRD4
- BRD7
- BRD9
- BRDT
References
- ↑ 1.0 1.1 PDB: 1e6i; "The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p". EMBO J. 19 (22): 6141–9. November 2000. doi:10.1093/emboj/19.22.6141. PMID 11080160.
- ↑ 2.0 2.1 2.2 Ntranos, Achilles; Casaccia, Patrizia (2016). "Bromodomains: Translating the words of lysine acetylation into myelin injury and repair". Neuroscience Letters 625: 4–10. doi:10.1016/j.neulet.2015.10.015. PMID 26472704.
- ↑ Zeng L, Zhou MM (February 2002). "Bromodomain: an acetyl-lysine binding domain". FEBS Lett. 513 (1): 124–8. doi:10.1016/S0014-5793(01)03309-9. PMID 11911891.
- ↑ "brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2". Cell 68 (3): 561–72. February 1992. doi:10.1016/0092-8674(92)90191-E. PMID 1346755.
- ↑ Ren, C; Zeng, L; Zhou, MM (2016). Preparation, Biochemical Analysis, and Structure Determination of the Bromodomain, an Acetyl-Lysine Binding Domain.. Methods in Enzymology. 573. pp. 321–43. doi:10.1016/bs.mie.2016.01.018. ISBN 9780128053652.
- ↑ Taniguchi, Yasushi (2016-11-07). "The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins". International Journal of Molecular Sciences 17 (11): 1849. doi:10.3390/ijms17111849. ISSN 1422-0067. PMID 27827996.
- ↑ Filippakopoulos, Panagis (2012). "Histone Recognition and Large-Scale Structural Analysis of the Human Bromodomain Family". Cell 149 (1): 214–231. doi:10.1016/j.cell.2012.02.013. PMID 22464331.
- ↑ Dhalluin, C; Carlson, J. E.; Zeng, L; He, C; Aggarwal, A. K.; Zhou, M. M.; Zhou, Ming-Ming (1999). "Structure and ligand of a histone acetyltransferase bromodomain". Nature 399 (6735): 491–6. doi:10.1038/20974. PMID 10365964.
- ↑ Santillan, D. A.; Theisler, C. M.; Ryan, A. S.; Popovic, R; Stuart, T; Zhou, M. M.; Alkan, S; Zeleznik-Le, N. J. (2006). "Bromodomain and histone acetyltransferase domain specificities control mixed lineage leukemia phenotype". Cancer Research 66 (20): 10032–9. doi:10.1158/0008-5472.CAN-06-2597. PMID 17047066.
- ↑ Hay, D. A.; Fedorov, O; Martin, S; Singleton, D. C.; Tallant, C; Wells, C; Picaud, S; Philpott, M et al. (2014). "Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains". Journal of the American Chemical Society 136 (26): 9308–19. doi:10.1021/ja412434f. PMID 24946055.
- ↑ Wang, Nian; Wu, Runliu; Tang, Daolin; Kang, Rui (2021-01-19). "The BET family in immunity and disease" (in en). Signal Transduction and Targeted Therapy 6 (1): 23. doi:10.1038/s41392-020-00384-4. ISSN 2059-3635. PMID 33462181.
- ↑ Jung, Marie; Gelato, Kathy A; Fernández-Montalván, Amaury; Siegel, Stephan; Haendler, Bernard (2015-06-16). "Targeting BET bromodomains for cancer treatment". Epigenomics 7 (3): 487–501. doi:10.2217/epi.14.91. PMID 26077433.
- ↑ Da Costa, D.; Agathanggelou, A.; Perry, T.; Weston, V.; Petermann, E.; Zlatanou, A.; Oldreive, C.; Wei, W. et al. (2013-07-19). "BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia" (in en). Blood Cancer Journal 3 (7): e126. doi:10.1038/bcj.2013.24. PMID 23872705.
- ↑ Gacias, Mar; Gerona-Navarro, Guillermo; Plotnikov, Alexander N.; Zhang, Guangtao; Zeng, Lei; Kaur, Jasbir; Moy, Gregory; Rusinova, Elena et al. (2014). "Selective Chemical Modulation of Gene Transcription Favors Oligodendrocyte Lineage Progression". Chemistry & Biology 21 (7): 841–854. doi:10.1016/j.chembiol.2014.05.009. ISSN 1074-5521. PMID 24954007.
- ↑ Shi, Junwei (2014). "The Mechanisms behind the Therapeutic Activity of BET Bromodomain Inhibition". Molecular Cell 54 (5): 728–736. doi:10.1016/j.molcel.2014.05.016. PMID 24905006.
- ↑ Clark, P. G.; Vieira, L. C.; Tallant, C; Fedorov, O; Singleton, D. C.; Rogers, C. M.; Monteiro, O. P.; Bennett, J. M. et al. (2015). "LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor". Angewandte Chemie International Edition 54 (21): 6217–21. doi:10.1002/anie.201501394. PMID 25864491.
- ↑ Theodoulou, N. H.; Bamborough, P; Bannister, A. J.; Becher, I; Bit, R. A.; Che, K. H.; Chung, C. W.; Dittmann, A et al. (2015). "The Discovery of I-BRD9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition". Journal of Medicinal Chemistry 59 (4): 1425–39. doi:10.1021/acs.jmedchem.5b00256. PMID 25856009.
 |
