Biology:C16orf78
Uncharacterized protein C16orf78(NP_653203.1) is a protein that in humans is encoded by the chromosome 16 open reading frame 78 gene.[1]
Gene
The C16orf78 gene(123970) is located at 16q12.1 on the plus strand, spanning 25,609 bp from 49,407,734 to 49,433,342.[2]
mRNA
There is one mRNA transcript (NM_144602.3) and no other known splice isoforms. There are 5 exons, totaling a length of 1068 base pairs.[2]
Protein
Sequence
C16orf78 is 265 amino acids long with a predicted molecular weight of 30.8 kDal and pI of 9.8.[3] It is rich in both methionine and lysine, composed of 6.4% methionine and 13.6% lysine.[4] This methionine richness has been hypothesized to serve as a mitochondrial antioxidant.[5]
Post-Transnational Modifications
There are four verified ubiquitination sites and three verified phosphorylation sites.[6][7]

Structure
Predictions of C16orf78's secondary structure consist primarily of alpha helices and coiled coils.[9][10][11] Phyre2 also predicted C16orf78 is primarily helical, but 253 of 265 amino acids were modeled ab initio so the confidence of the model is low.[12]
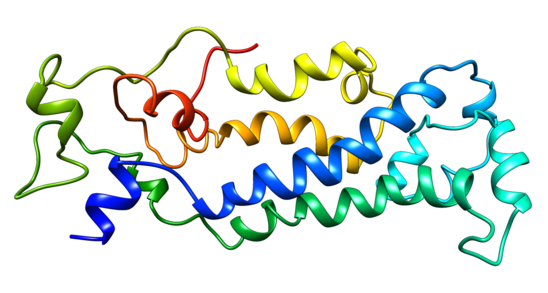
Subcellular Localization
C16orf78 is predicted to be localized to the cell nucleus.[13] There is also a predicted bipartite nuclear localization signal.[14]
Expression
C16orf78 has restricted expression toward the testis, with much lower expression in other tissues.[15]

Interaction
C16orf78 has a physical association with DNA/RNA-binding protein KIN17 (NP_036443.1), suggesting C16orf78 may also play a role in DNA repair.[17] C16orf78 was found to be phosphorylated by SRPK1(NP_003128.3) and SPRK2 (AAH68547.1).[6]
Clinical Significance
Deletion of the C16orf78 gene has been identified as a determinant of prostate cancer.[18] A SNP in C16orf78 interacts with a SNP in LMTK2 and is associated with risk of prostate cancer.[19]
Amplification of the C16orf78 gene has been linked to metabolically adaptive cancer cells.[20] A duplication of the C16orf78 gene was associated with at least one case of Rolandic Epilepsy.[21]
Homology
Paralogs
C16orf78 has no known paralogs in humans.[22]
Orthologs
C16orf78 has over 80 orthologs, including animals as distant Ciona intestinalis(XP_002132057.1), which is estimated to have diverged from humans 676 million years ago.[2][23] C16orf78 has orthologs in many types of mammals, reptiles, bony fish, and even some invertebrates, but has no known orthologs in amphibians or birds.[22] Below is a table with samples of orthologs, with divergence dates from TimeTree and similarity calculated by pairwise sequence alignment.[24]
| Species Name | NCBI Accession | Divergence (mya) (estimated) | Length (aa) | % Identity | % Similarity |
| Homo sapiens | NP_653203.1 | 0 | 265 | 100% | 100% |
| Gorilla gorilla gorilla | XP_004057673.2 | 9.06 | 265 | 96% | 98% |
| Macaca mulatta | XP_001082258.1 | 29.44 | 267 | 89% | 93% |
| Galeopterus variegatus | XP_008591134.1 | 76 | 266 | 65% | 77% |
| Oryctolagus cuniculus | XP_008273281.1 | 90 | 255 | 62% | 76% |
| Mus musculus | NP_808569.1 | 90 | 270 | 57% | 69% |
| Lipotes vexillifer | XP_007459548.1 | 96 | 266 | 65% | 77% |
| Capra hircus | XP_017918754.1 | 96 | 276 | 63% | 74% |
| Callorhinus ursinus | XP_025708226.1 | 96 | 250 | 62% | 74% |
| Pteropus vampyrus | XP_011358492.1 | 96 | 263 | 60% | 74% |
| Loxodonta africana | XP_023411324.1 | 105 | 285 | 48% | 55% |
| Sarcophilus harrisii | XP_003757266.1 | 159 | 270 | 38% | 53% |
| Vombatus ursinus | XP_027723426.1 | 159 | 275 | 38% | 54% |
| Pogona vitticeps | XP_020643996.1 | 312 | 315 | 26% | 43% |
| Gekko japonicus | XP_015263322.1 | 312 | 261 | 25% | 47% |
| Python bivittatus | XP_025030465.1 | 312 | 313 | 23% | 37% |
| Latimeria chalumnae | XP_014344069.1 | 413 | 310 | 19% | 42% |
| Acipenser ruthenus | RXM34621.1 | 435 | 202 | 15% | 37% |
| Ciona intestinalis | XP_002132057.1 | 676 | 396 | 10% | 32% |
| Apostichopus japonicus | PIK46940.1 | 684 | 292 | 9% | 33% |
References
- ↑ "uncharacterized protein C16orf78 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_653203.1.
- ↑ 2.0 2.1 2.2 "Gene: C16orf78 (ENSG00000166152) - Summary - Homo sapiens - Ensembl genome browser 96". http://useast.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000166152;r=16:49373823-49399431;t=ENST00000299191.
- ↑ "ExPASy - ProtParam tool". https://web.expasy.org/protparam/.
- ↑ "SAPS < Sequence Statistics < EMBL-EBI". https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ Schindeldecker, Mario; Moosmann, Bernd (10 April 2015). "Protein-borne methionine residues as structural antioxidants in mitochondria". Amino Acids 47 (7): 1421–1432. doi:10.1007/s00726-015-1955-8. PMID 25859649.
- ↑ 6.0 6.1 "C16orf78 Result Summary | BioGRID". https://thebiogrid.org/125845/summary/homo-sapiens/c16orf78.html.
- ↑ "C16orf78 (human)". https://www.phosphosite.org/proteinAction?id=16329&showAllSites=true.
- ↑ "PROSITE". https://prosite.expasy.org/cgi-bin/prosite/mydomains/.
- ↑ "CFSSP: Chou & Fasman Secondary Structure Prediction Server". http://www.biogem.org/tool/chou-fasman/.
- ↑ "NPS@ : GOR4 secondary structure prediction". https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html.
- ↑ "JPred: A Protein Secondary Structure Prediction Server" (in en). http://www.compbio.dundee.ac.uk/jpred/.
- ↑ Kelley, Lawrence A; Mezulis, Stefans; Yates, Christopher M; Wass, Mark N; Sternberg, Michael J E (7 May 2015). "The Phyre2 web portal for protein modeling, prediction and analysis". Nature Protocols 10 (6): 845–858. doi:10.1038/nprot.2015.053. PMID 25950237.
- ↑ Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. (8 May 2007). "WoLF PSORT: protein localization predictor". Nucleic Acids Research 35 (Web Server): W585–W587. doi:10.1093/nar/gkm259. PMID 17517783.
- ↑ "Motif Scan" (in en). https://myhits.isb-sib.ch/cgi-bin/motif_scan.
- ↑ "C16orf78 chromosome 16 open reading frame 78 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&list_uids=123970#gene-expression.
- ↑ "49000288 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/49000288.
- ↑ IntAct. "IntAct Portal" (in en). https://www.ebi.ac.uk/intact/interaction/EBI-20903736.
- ↑ DePihno, R. A et al. (2016). U.S. Patent No. 9458510. Washington, DC: U.S. Patent and Trademark Office.
- ↑ Tao, Sha; Wang, Zhong; Feng, Junjie; Hsu, Fang-Chi; Jin, Guangfu; Kim, Seong-Tae; Zhang, Zheng; Gronberg, Henrik et al. (March 2012). "A genome-wide search for loci interacting with known prostate cancer risk-associated genetic variants". Carcinogenesis 33 (3): 598–603. doi:10.1093/carcin/bgr316. PMID 22219177.
- ↑ Singh, Balraj; Shamsnia, Anna; Raythatha, Milan R.; Milligan, Ryan D.; Cady, Amanda M.; Madan, Simran; Lucci, Anthony; Das, Gokul M. (3 October 2014). "Highly Adaptable Triple-Negative Breast Cancer Cells as a Functional Model for Testing Anticancer Agents". PLOS ONE 9 (10): e109487. doi:10.1371/journal.pone.0109487. PMID 25279830. Bibcode: 2014PLoSO...9j9487S.
- ↑ Reinthaler, Eva M.; Lal, Dennis; Lebon, Sebastien; Hildebrand, Michael S.; Dahl, Hans-Henrik M.; Regan, Brigid M.; Feucht, Martha; Steinböck, Hannelore et al. (15 November 2014). "16p11.2 600 kb Duplications confer risk for typical and atypical Rolandic epilepsy". Human Molecular Genetics 23 (22): 6069–6080. doi:10.1093/hmg/ddu306. PMID 24939913.
- ↑ 22.0 22.1 "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "TimeTree :: The Timescale of Life". http://www.timetree.org/.
- ↑ "Pairwise Sequence Alignment Tools < EMBL-EBI". https://www.ebi.ac.uk/Tools/psa/.
External links
- Human C16orf78 genome location and C16orf78 gene details page in the UCSC Genome Browser.
 |

