Biology:CIMAP1C
CIMAP1C (ciliary microtubule associated protein 1C) is a gene in humans that encodes the CIMAP1C protein. It is also often referred to as ODF3L1 (outer dense fiber protein 3-like protein 1). CIMAP1C is expressed in low levels throughout the body with high expression levels in the testes. It is highly conserved in mammals and reptiles but not present in birds or amphibians, indicating it arose around 300 million years ago.
Gene
The CIMAP1C gene is found on Chromosome 15 at position 15q24.2, spanning 3.72 kb.[1] It contains 4 exons and 1,132 nucleotides.[2]
Aliases
CIMAP1C is also known as ODF3L1 (outer dense fiber protein 3 like protein 1).[2]
Protein
The CIMAP1C protein is 274 amino acids long and its molecular weight is ~31 kDa.[4] Its isoelectric point is 9.6.[5] There are no known isoforms.[2] CIMAP1C is very basic, proline rich, and highly tyrosine rich.[6] There are two structurally significant regions known as sperm tail proline-glycine rich repeat regions.[2] These regions are highly conserved and contain glycosylation and phosphorylation sites.[7]
Gene level regulation
CIMAP1C is expressed at low levels in most tissues in the body (15.7%), with high expression in the testis.[2] Slightly high levels of expression are seen in the mammary glands.[2]
Protein level regulation
CIMAP1C is imported into the nucleus via two nuclear localization signals, and can otherwise be found in the cytoskeleton.[2][8] CIMAP1C contains a variety of phosphorylation sites, specifically protein kinase C and Casein kinase II phosphorylation.[7]
Evolution
Paralogs
There are no paralogs of the CIMAP1C gene.[2]
Orthologs
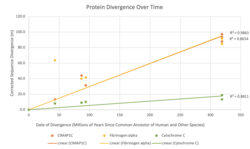
CIMAP1C is highly conserved, and can be found in mammals and reptiles. CIMAP1C is not found in amphibians or birds, therefore it arose 300 million years ago.[9] Based on the calculated protein divergence figure, CIMAP1C evolves rapidly and similarly to Fibrinogen Alpha. The table below lists orthologs and their information.
| Genus and Species | Common Name | Taxonomic Group | Mediat Date of Divergence (MYA)[9] | Accession number | Sequence length (aa) | Sequence Identity to human protein (%) | Sequence Similarity to human Protein (%) |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | Primates | 0.0 | NP_787077.1 | 274 | 100.0 | 100.0 |
| Saimiri boliviensis | Squirrel Monkey | Primates | 43 | XP_010329148.1 | 274 | 87.6 | 92.7 |
| Canis lupus familiaris | Dog | Carnivora | 94 | XP_038297715.1 | 275 | 78.2 | 88.4 |
| Mus musculus | House Mouse | Rodentia | 87 | NP_001361610.1 | 306 | 64.4 | 75.5 |
| Myotis brandtii | Brandt's Bat | Chiroptera | 94 | XP_005884580.1 | 275 | 74.5 | 85.8 |
| Gracilinanus agilis | Mouse Opossum | Didelphimorphia | 160 | XP_044518760.1 | 278 | 58.8 | 73.8 |
| Bubalus bubalis | Water buffalo | Artiodactyla | 94 | XP_006080752.3 | 274 | 73.1 | 84.4 |
| Pogona vitticeps | Cental Bearded Dragon | Squamata | 319 | XP_020640486.1 | 285 | 40.2 | 54.3 |
| Notechis scutatus | Tiger Snake | Squamata | 319 | XP_026540911.1 | 195 | 28.4 | 41.4 |
| Dermochelys coriacea | Leatherback Sea Turtle | Testudines | 319 | XP_038274105.1 | 274 | 37.8 | 54.7 |
| Alligator mississippiensis | American Alligator | Crocodylia | 319 | XP_019333216.1 | 249 | 37.8 | 52.8 |
| Phascolarctos cinereus | Koala | Diprotodontia | 160 | XP_020850090.1 | 278 | 59 | 73.4 |
| Phocoena sinus | Vaquita | Artiodactyla | 94 | XP_032479987.1 | 275 | 77.5 | 87.3 |
| Eublepharis macularius | Leopard gecko | Squamata | 319 | XP_054858185.1 | 276 | 38.5 | 50.7 |
| Ornithorhynchus anatinus | Platypus | Monotremata | 180 | XP_001509549.1 | 269 | 45.8 | 58.8 |
Interacting proteins
It has been experimentally determined that CIMAP1C interacts with STAMBP (STAM binding protein) through affinity chromatography.[10] STAMBP regulates cell surface receptor-mediated endocytosis and ubiquitin-dependent receptor sorting to lysosomes.[11]
Clinical significance
Expression levels of CIMAP1C are positive markers of disease prognosis for those with papillary thyroid carcinoma (PTC) and non-metastatic breast cancer (NMBC). High levels of CIMAP1C in tumor tissues accurately predicted a better prognosis for individuals with PTC.[12] CIMAP1C is also noted as a positive indicator for the effectiveness of NMBC radiotherapy.[13]
References
- ↑ "AceView: a comprehensive cDNA-supported gene and transcripts annotation". 11 June 2023. https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 "CIMAP1C ciliary microtubule associated protein 1C [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/161753.
- ↑ Zhang, Yang (2009). "I-TASSER: Fully automated protein structure prediction in CASP8". Proteins: Structure, Function, and Bioinformatics 77 (S9): 100–113. doi:10.1002/prot.22588. ISSN 0887-3585. PMID 19768687. PMC 2782770. http://dx.doi.org/10.1002/prot.22588.
- ↑ "Thermo Fisher ODF3L1 Antibodies - polyclonal rabbit anti-human ODF3L1 antibody". 9 July 2023. https://www.thermofisher.com/antibody/product/ODF3L1-Antibody-Polyclonal/PA5-69939.
- ↑ Duvaud, Séverine; Gabella, Chiara; Lisacek, Frédérique; Stockinger, Heinz; Ioannidis, Vassilios; Durinx, Christine (2021-04-13). "Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users". Nucleic Acids Research 49 (W1): W216–W227. doi:10.1093/nar/gkab225. ISSN 0305-1048. PMID 33849055. PMC 8265094. http://dx.doi.org/10.1093/nar/gkab225.
- ↑ "EMBL-EBI Tools: An introduction". EMBL's European Bioinformatics Institute. doi:10.6019/tol.ebitools-t.2021.00001.1. http://dx.doi.org/10.6019/tol.ebitools-t.2021.00001.1.
- ↑ 7.0 7.1 Blom, Nikolaj; Gammeltoft, Steen; Brunak, Søren (December 1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites". Journal of Molecular Biology 294 (5): 1351–1362. doi:10.1006/jmbi.1999.3310. ISSN 0022-2836. http://dx.doi.org/10.1006/jmbi.1999.3310.
- ↑ Thumuluri, Vineet; Almagro Armenteros, José Juan; Rosenberg Johansen, Alexander; Nielsen, Henrik; Winther, Ole (2022-04-30). "DeepLoc 2.0: multi-label subcellular localization prediction using protein language models". Nucleic Acids Research 50 (W1): W228–W234. doi:10.1093/nar/gkac278. ISSN 0305-1048. PMID 35489069. PMC 9252801. http://dx.doi.org/10.1093/nar/gkac278.
- ↑ 9.0 9.1 "Data S1: RaxML file, TimeTree, and alignments". doi:10.7717/peerj.5401/supp-3.
- ↑ "ODF3L1 protein (human) - STRING interaction network". https://string-db.org/cgi/network?taskId=bqrBLEHavKZ3&sessionId=bn6f1Fl7aZt5.
- ↑ Huttlin, Edward L.; Bruckner, Raphael J.; Navarrete-Perea, Jose; Cannon, Joe R.; Baltier, Kurt; Gebreab, Fana; Gygi, Melanie P.; Thornock, Alexandra et al. (May 2021). "Dual proteome-scale networks reveal cell-specific remodeling of the human interactome". Cell 184 (11): 3022–3040.e28. doi:10.1016/j.cell.2021.04.011. ISSN 0092-8674. PMID 33961781. PMC 8165030. http://dx.doi.org/10.1016/j.cell.2021.04.011.
- ↑ Guo, Mengli; Chen, Zhen; Li, Yayi; Li, Sijin; Shen, Fei; Gan, Xiaoxiong; Feng, Jianhua; Cai, Wensong et al. (2021-06-23). "Tumor Mutation Burden Predicts Relapse in Papillary Thyroid Carcinoma With Changes in Genes and Immune Microenvironment". Frontiers in Endocrinology 12. doi:10.3389/fendo.2021.674616. ISSN 1664-2392. PMID 34248843.
- ↑ Chen, Huajian; Huang, Li; Wan, Xinlong; Ren, Shigang; Chen, Haibin; Ma, Shumei; Liu, Xiaodong (March 2023). "Polygenic risk score for prediction of radiotherapy efficacy and radiosensitivity in patients with non-metastatic breast cancer" (in en). Radiation Medicine and Protection 4 (1): 33–42. doi:10.1016/j.radmp.2023.01.001.
 |
