Biology:Catechol 1,2-dioxygenase
| Catechol 1,2-Dioxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.13.11.1 | ||||||||
| CAS number | 9027-16-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
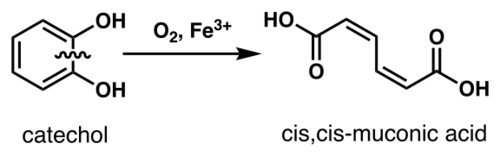
Catechol 1,2- dioxygenase (EC 1.13.11.1, 1,2-CTD, catechol-oxygen 1,2-oxidoreductase, 1,2-pyrocatechase, catechase, catechol 1,2-oxygenase, catechol dioxygenase, pyrocatechase, pyrocatechol 1,2-dioxygenase, CD I, CD II)[1][2][3][4] is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:[5]
More specifically, 1,2-CTD is an intradiol dioxygenase, a family of catechol dioxygenases that cleaves the bond between the phenolic hydroxyl groups of catechol using an Fe3+ cofactor.
Thus far, 1,2-CTD has been observed to exist in the following species of soil bacteria and fungi: Pseudomonas sp.,[6] Pseudomonas fluorescens,[7] Aspergillus niger,[8] Brevibacterium fuscum,[9][10] Acinetobacter calcoaceticus,[11] Trichosporon cutaneum,[12] Rhodococcus erythropolis,[13] Frateuria sp.,[14] Rhizobium trifolii,[15] Pseudomonas putida,[16] Candida tropicalis,[17] Candida maltose,[18] Rhizobium leguminosarum,[19] and Nocardia sp..[20] These bacteria subsequently employ 1,2-CTD in the last step of the degradation of aromatic compounds to aliphatic products.[21]
History
Two families of dioxygenases were discovered by Osamu Hayaishi and Kizo Hashimoto in 1950: catechol 1,2-dioxygenase and catechol 2,3-dioxygenase (2,3-CTD).[22] The two enzymes were identified to be a part of two separate catechol dioxygenase families: 1,2-CTD was classified as an intradiol dioxygenase while 2,3-CTD was classified as an extradiol dioxygenase. The two enzymes can be distinguished based on their reaction products and cofactors. 1,2-CTD uses Fe3+ as a cofactor to cleave the carbon-carbon bond between the phenolic hydroxyl groups of catechol, thus yielding muconic acid as its product. In contrast, 2,3-CTD utilizes Fe2+ as a cofactor to cleave the carbon-carbon bond adjacent to the phenolic hydroxyl groups of catechol, thus yielding 2-hydroxymuconaldehye as its product.[23]
Enzyme Structure
Almost all members of the 1,2-CTD family are homodimers; the 1,2-CTD enzyme produced by Pseudomonas arvilla is the exception to this rule, containing two highly homologous subunits that can form either a homo- or hetero- dimer.[24] The enzyme resembles a boomerang in shape, and can therefore be clearly divided into three domains: two catalytic domains residing at each end of the “boomerang” and a linker domain at the center.
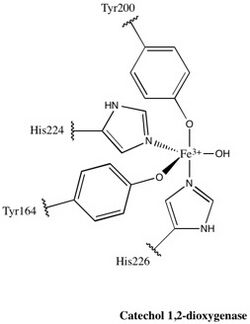
Each catalytic domain is composed of two stacked, mixed topology β sheets and several random coils. These sheets and coils subsequently encompass the active site: a non-heme iron(III) complex. Without heme, iron must be ligated to four amino acid residues (Tyr200, His226, Tyr164, His224) to maintain is catalytically active conformation. With Tyr200 and His226 acting as the axial ligands and Tyr164, His224, and a solvent water molecule acting as equatorial ligands, the Fe3+ complex displays trigonal bipyramidal geometry. Since the active sites of each catalytic domain are separated by a distance of 40 Å, they are not believed to allosterically effect one another.
In contrast, the linker domain is composed of α helices supplied by the two catalytic domains: each domain contributes five helices from their N termini and one from a helix that spans both the catalytic domain and the linker domain. At the center of the linker domain resides an 8 by 35 Å hydrophobic tunnel with two phospholipids bound at each end. The head of each phospholipid points outward towards solution while the tails are embedded within the enzyme. The function of this hydrophobic tunnel is unknown, though two hypotheses have been postulated concerning its utility. The first is that the binding of the terminal phospholipids alters the conformation of the active sites, implying that the tunnel acts as an effector, only allowing the enzyme to be active in certain areas of the cell. The second hypothesis postulates that the tunnel regulates lipid membrane rigidity through its degradation of phenolic hydrocarbons and ability to bind to other lipids. Studies have shown that phenolic hydrocarbons affect the functional and structural properties of cell membranes.[25] 1,2-CTD degrades phenolic hydrocarbons key to the synthesis lipid membranes. Therefore, 1,2-CTD may bind to the cell lipid membrane via its terminal phospholipids and thus have greater access to the phenolic hydrocarbons vital in lipid membrane structure.[26]
Enzyme Mechanism
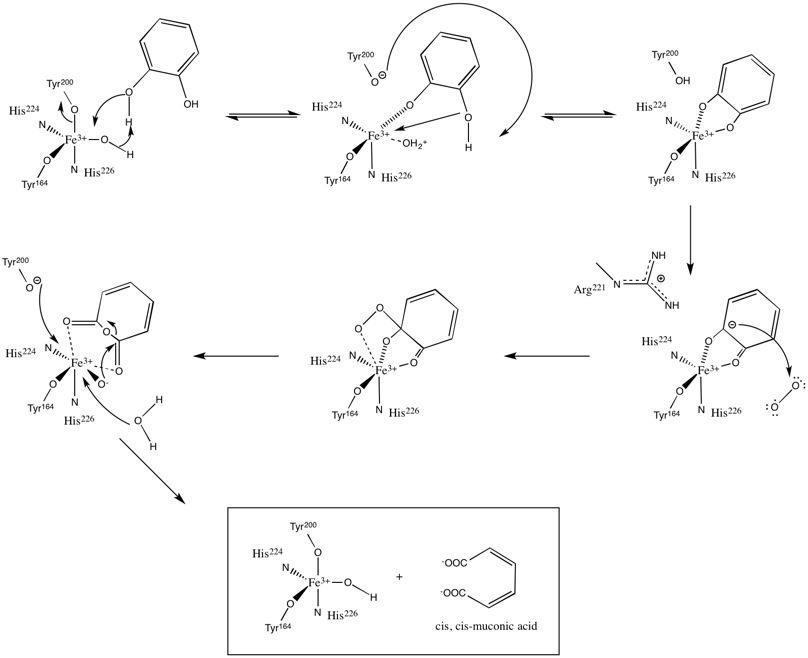
The catalytic mechanism of catechol 1,2-dioxygenase was elucidated using a combination of O18 labeling experiments and crystallography.[27][28][29] Upon entering the active site, the hydroxyl group on the fourth carbon (C4) of catechol binds to Fe3+; this binding is facilitated by the hydroxide ligand, which deprotonates the C4 hydroxyl group. The second catechol hydroxyl group on carbon 3 (C3) is coordinated to Fe3+ after its deprotonation by the Tyr200 ligand. In the process of deprotonating the C3 hydroxyl group, Tyr200 dissociates from the catalytic complex.
Oxygen is bonded to the substrate through a series of trans influences and stabilizing hydrogen bonding between the substrate and other active site amino acid residues. His226 accepts electron density from the substrate, consequently decreasing the bond between Fe3+ and the C4 hydroxyl. At the same time the bond between the C3 hydroxyl and Fe3+ is increased due to the electron withdrawing effects of Tyr164. These distortions, coupled with the hydrogen bonding between Arg221 and the C3 hydroxyl, induces the C3 hydroxyl group to ketonize and increases the carbanion character of C4. The newly formed C4 carbanion attacks O2, thus binding it to the substrate.
Another trans influence follows the aforementioned step, leading to the cleavage of O2 and the subsequent insertion of one of the oxygen molecules between C3 and C4. Rapid hydrolysis follows this reaction, yielding a primed active site and product.
References
- ↑ "Direct oxygenation by O2, oxygenases". The Enzymes. 8 (2nd ed.). New York: Academic Press. 1963. pp. 353–371.
- ↑ "Studies on oxygenases; pyrocatechase". The Journal of Biological Chemistry 229 (2): 905–20. December 1957. PMID 13502352.
- ↑ "The mechanism of formation of beta-ketoadipic acid by bacteria". The Journal of Biological Chemistry 210 (2): 821–36. October 1954. PMID 13211620.
- ↑ "Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2". Applied and Environmental Microbiology 52 (2): 334–9. August 1986. doi:10.1128/AEM.52.2.334-339.1986. PMID 3752997.
- ↑ Schomburg, Dietmar; Schomburg, Ida (2006). "Catechol 1,2-dioxygenase". Class I Oxidoreductases X. Springer Handbook of Enzymes. 25 (2nd ed.). Springer Berlin Heidelberg. pp. 382–394. doi:10.1007/3-540-37704-2_47. ISBN 978-3-540-37704-7. https://www.springer.com/life+sciences/biochemistry+%26+biophysics/book/978-3-540-26585-6.
- ↑ "Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad". The Biochemical Journal 174 (1): 73–84. July 1978. doi:10.1042/bj1740073. PMID 697765.
- ↑ "Pioneering the Field of Oxygenases through the Study of Tryptophan Metabolism: the Work of Osamu Hayaishi". J. Biol. Chem. 229: 905–920. 1957. http://www.jbc.org/content/280/47/e44.full.pdf.
- ↑ "Catechol 1,2-dioxygenase from Aspergillus niger: Purification and properties". J. Indian Inst. Sci. 63C: 131–136. 1981. http://journal.iisc.ernet.in/index.php/iisc/article/viewFile/3871/3906.
- ↑ "Characteristics of Catechol Oxygenase from Brevibacterium fuscum". J. Biochem. 54 (1): 65–74. 1963. doi:10.1093/oxfordjournals.jbchem.a127748. PMID 14056355.
- ↑ "Extradiol cleavage of 3-methylcatechol by catechol 1,2-dioxygenase from various microorganisms". Applied and Environmental Microbiology 33 (3): 725–7. March 1977. doi:10.1128/AEM.33.3.725-727.1977. PMID 16345232.
- ↑ "Catechol 1,2-dioxygenase from Acinetobacter calcoaceticus: purification and properties". Journal of Bacteriology 127 (1): 536–44. July 1976. doi:10.1128/JB.127.1.536-544.1976. PMID 58860.
- ↑ Itoh, M (1981). "Characteristics of a new catechol-1,2-oxygenase from Trichosporon cutaneum WY2-2". Agric. Biol. Chem. 45 (1): 2787–2796.
- ↑ "Classification of catechol 1,2-dioxygenase family: sequence analysis of a gene for the catechol 1,2-dioxygenase showing high specificity for methylcatechols from Gram+ aniline-assimilating Rhodococcus erythropolis AN-13". Agric. Biol. Chem. 48 (1): 2087–2095. 1984. doi:10.1016/S0378-1119(96)00629-4. PMID 9034312.
- ↑ "Two catechol 1,2-dioxygenases from aniline-assimilating bacterium, Frateuria species ANA-18". Agric. Biol. Chem. 48 (1): 2097–2104. 1984.
- ↑ "Aromatic metabolism in Rhizobium trifolii-catechol 1,2-dioxygenase". Arch. Microbiol. 141 (1): 225–228. 1985. doi:10.1007/BF00408063.
- ↑ "Reactions of 3-ethylcatechol and 3-(methylthio)catechol with catechol dioxygenases". Archives of Biochemistry and Biophysics 248 (1): 130–7. July 1986. doi:10.1016/0003-9861(86)90409-1. PMID 3015028.
- ↑ "Degradation of phenolic compounds by the yeast Candida tropicalis HP 15. II. Some properties of the first two enzymes of the degradation pathway". Journal of Basic Microbiology 26 (5): 271–81. 1986. doi:10.1002/jobm.3620260505. PMID 3783431.
- ↑ "Purification and characterization of pyrocatechase from the catechol- assimilating yeast Candida maltose". Agric. Biol. Chem. 52 (2): 585–587. 1988. doi:10.1271/bbb1961.52.585.
- ↑ "Purification and Properties of Catechol 1,2-Dioxygenase from Rhizobium leguminosarum biovar viceae USDA 2370". Applied and Environmental Microbiology 56 (6): 1971–3. June 1990. doi:10.1128/AEM.56.6.1971-1973.1990. PMID 16348234.
- ↑ "Properties of cyanogen bromide-activated, Agarose-immobilized catechol 1,2-dioxygenase from freeze-dried extracts of Nocardia sp. NCIB 10503". Enzyme Microb. Technol. 12 (12): 945–949. 1990. doi:10.1016/0141-0229(90)90114-6.
- ↑ "Dioxygen Activation by Enzymes with Mononuclear Non-Heme Iron Active Sites". Chemical Reviews 96 (7): 2607–2624. November 1996. doi:10.1021/cr960039f. PMID 11848838.
- ↑ Hayaishi, Osamu; Hashimoto, Kizo (1950). "Pyrocatecase a New Enzyme Catalizing Oxidative Breakdown of Pyrocatechin". J. Biochem. 37 (3): 371–374. doi:10.1093/oxfordjournals.jbchem.a126205.
- ↑ Bugg, Timothy (2003). "Dioxygenase enzymes: catalytic mechanisms and chemical models". Tetrahedron 59 (36): 7075–7101. doi:10.1016/S0040-4020(03)00944-X.
- ↑ "Nonidentical subunits of pyrocatechase from Pseudomonas arvilla C-1". Archives of Biochemistry and Biophysics 195 (1): 12–22. June 1979. doi:10.1016/0003-9861(79)90322-9. PMID 475379.
- ↑ "Interactions of cyclic hydrocarbons with biological membranes". The Journal of Biological Chemistry 269 (11): 8022–8. March 1994. PMID 8132524. http://gbb.eldoc.ub.rug.nl/FILES/root/1994/JBiolChemSikkema/1994JBiolChemSikkema.pdf?origin=publication_detail.[yes|permanent dead link|dead link}}]
- ↑ "The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker". Structure 8 (4): 429–40. April 2000. doi:10.1016/S0969-2126(00)00122-2. PMID 10801478.
- ↑ Hayaishi, Osamu; Hashimoto, Kizo (1950). "Pyrocatecase a New Enzyme Catalizing Oxidative Breakdown of Pyrocatechin". J. Biochem. 37 (3): 371–374. doi:10.1093/oxfordjournals.jbchem.a126205.
- ↑ "Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding". Biochemistry 36 (33): 10052–66. August 1997. doi:10.1021/bi970469f. PMID 9254600.
- ↑ Metal Ions in Biological Systems. 28. Marcel Dekker, Inc.. 1992. pp. 243–298.
External links
- Catechol+1,2-dioxygenase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |