Biology:CrfA RNA
| CrfA RNA | |
|---|---|
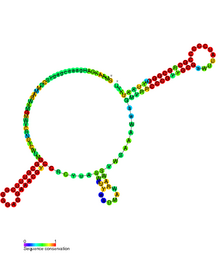 Conserved secondary structure of CrfA RNA. | |
| Identifiers | |
| Symbol | CrfA |
| Other data | |
| Domain(s) | Caulobacter |
| PDB structures | PDBe |
CrfA RNA (Caulobacter response to famine RNA) is a family of non-coding RNAs found in Caulobacter crescentus. CrfA is expressed upon carbon starvation and is thought to activate 27 genes.[1] It was originally identified along with 26 other non-coding RNAs using a tiled Caulobacter microarray protocol specifically aimed at detecting small RNAs.[2]
CrfA RNA is one of only 8 putative ncRNAs conserved in the closely related Caulobacter sp. K31.[1]
Carbon starvation response
CrfA was found to be upregulated 10-fold in C. crescentus when glucose was deprived in a minimal medium. Further experimentation found the response to be specific to carbon deprivation; its expression was not increased when phosphate or nitrogen were limiting factors.[1]
Affymetrix microarrays were then used to analyse changes in the transcriptome in response to increased CrfA. Seven affected gene products were TonB-dependent receptors, outer membrane proteins which facilitate the uptake of external substrate.[3] The upregulation of these proteins could increase the carbon uptake during starvation. Another CrfA regulated gene (EntrezGene CC_1363) is thought to encode a proton pump powered by pyrophosphate hydrolysis. Increasing production of this protein could enable the cell to maintain its electrochemical gradient and power ATP synthesis during carbon starvation.[1]
σ54 regulates many other proteins involved in the carbon starvation response at the level of transcription.[4]
References
- ↑ 1.0 1.1 1.2 1.3 "CrfA, a Small Noncoding RNA Regulator of Adaptation to Carbon Starvation in Caulobacter crescentus". J. Bacteriol. 192 (18): 4763–4775. September 2010. doi:10.1128/JB.00343-10. PMID 20601471.
- ↑ "Small non-coding RNAs in Caulobacter crescentus". Mol. Microbiol. 68 (3): 600–614. May 2008. doi:10.1111/j.1365-2958.2008.06172.x. PMID 18373523.
- ↑ Koebnik R (August 2005). "TonB-dependent trans-envelope signalling: the exception or the rule?". Trends Microbiol. 13 (8): 343–347. doi:10.1016/j.tim.2005.06.005. PMID 15993072.
- ↑ "Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability". J. Bacteriol. 192 (3): 819–833. February 2010. doi:10.1128/JB.01240-09. PMID 19948804.
Further reading
- "Systems biology of Caulobacter". Annu. Rev. Genet. 41: 429–441. 2007. doi:10.1146/annurev.genet.41.110306.130346. PMID 18076330. http://pdfs.semanticscholar.org/ec34/bc7843e9c4042cbb29cd0390047a32abcaf6.pdf.
- "The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator". Mol. Microbiol. 47 (4): 929–941. February 2003. doi:10.1046/j.1365-2958.2003.03349.x. PMID 12581350.
- "Regulatory roles for small RNAs in bacteria". Curr. Opin. Microbiol. 6 (2): 120–124. April 2003. doi:10.1016/S1369-5274(03)00027-4. PMID 12732300. https://zenodo.org/record/1260240.
- "Bacterial small RNA regulators". Crit. Rev. Biochem. Mol. Biol. 40 (2): 93–113. 2005. doi:10.1080/10409230590918702. PMID 15814430. https://zenodo.org/record/1234471.
- "Genetics of Caulobacter crescentus". Bacterial Genetic Systems. Methods in Enzymology. 204. 1991. pp. 372–384. doi:10.1016/0076-6879(91)04019-K. ISBN 9780121821050.
- "Activation of gene expression by small RNA". Curr. Opin. Microbiol. 12 (6): 674–682. December 2009. doi:10.1016/j.mib.2009.09.009. PMID 19880344.
 |

