Biology:Haplogroup O-M175
| Haplogroup O-M175 | |
|---|---|
 | |
| Possible time of origin | 41,750 (95% CI 30,597<-> 46,041) years ago[1] 44,700 or 38,300 ybp[2] |
| Coalescence age | 33,943 (95% CI 25,124 <-> 37,631) years (Karmin 2022[1]) 35,000 or 30,000 years ago depending on mutation rate[2] |
| Possible place of origin | Southeast Asia or East Asia |
| Ancestor | NO |
| Descendants | Primary: O1 (O-F265); O2 (O-M122) Secondary: O1a (O-M119); O1b (O-M268); O2a (O-M324); O2b (O-F742) |
| Defining mutations | M175 (+ numerous other SNPs).[3] |
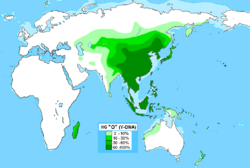
Haplogroup O, also known as O-M175, is a human Y-chromosome DNA haplogroup. It is primarily found among populations in Southeast Asia and East Asia. It also is found in various percentages of populations of the Russian Far East, South Asia, Central Asia, Caucasus, Crimea, Ukraine , Iran, Oceania, Madagascar and the Comoros. Haplogroup O is a primary descendant of haplogroup NO-M214.
The O-M175 haplogroup is very common amongst males from East and Southeast Asia. It has two primary branches: O1 (O-F265) and O2 (O-M122). O1 is found at high frequencies amongst males native to Southeast Asia, Taiwan, the Japan , the Korean Peninsula, Madagascar and some populations in southern China and Austroasiatic speakers of India . O2 is found at high levels amongst Han Chinese, Tibeto-Burman populations (including many of those in Yunnan, Tibet, Burma, Northeast India, and Nepal), Manchu, Mongols (especially those who are citizens of the PRC), Koreans, Vietnamese, Filipinos, Thais, Polynesians, Miao people, Hmong, the Naiman tribe of Kazakhs in Kazakhstan,[4] Kazakhs in the southeast of Altai Republic,[5] and Kazakhs in the Ili area of Xinjiang.[6]
Origins
Haplogroup O-M175 is a descendant haplogroup of Haplogroup NO-M214, and first appeared according to different theories either in Southeast Asia (see Rootsi 2006, TMC 1998, Shi 2005, and Bradshaw Foundation) or East Asia (see ISOGG 2012) approximately 40,000 years ago (or between 31,294 and 51,202 years ago according to Karmin et al. 2015).[7][8]
Haplogroup O-M175 is one of NO-M214's two branches. The other is Haplogroup N, which is common throughout North Eurasia.
Distribution
This haplogroup appears in high to moderate frequencies in most populations in both East Asia and Southeast Asia, and it is almost exclusive to that region: It is almost nonexistent in Western Siberia, Western Asia, Europe, most of Africa, India and the Americas, where its presence may be the result of recent migrations. However, certain O subclades do achieve significant frequencies among some populations of Central Asia, South Asia, and Oceania. For example, one study found it at a rate of 67% among the Naimans, a tribe in Kazakhstan,[4] even though the rate among Kazakhs in general is only about 3.3% to 10.8%(Wells et al. 2001).[4][9] (It is notable that 75% of cases of haplogroup O-M175 observed in the Kazakh sample of Ashirbekov et al. 2017, of which 10.8% have been found to belong to haplogroup O-M175, have been contributed by the Naimans themselves; only 3.1% of the remainder of the Kazakh sample with the Naimans excluded belong to haplogroup O-M175.) It has been estimated that 25% of the entire male population of the world carries different subclades of O.[8][10] Karafet et al. (2015) have assigned the Y-DNA of 46.2% (12/26) of a sample of Papuan from Pantar Island to haplogroup NO-M214;[11] considering their location in the Malay Archipelago, all or most of these individuals should belong to haplogroup O-M175.
An association with the spread of Austronesian languages in late antiquity is suggested by significant levels of O-M175 among island populations of the South Pacific and Indian Ocean, including the East African littoral. For example, Haplogroup O-M50 has even been found in Bantu-speaking populations of the Comoros along 6% of O-MSY2.2(xM50),[12] while both O-M50 and O-M95(xM88) occur commonly among the Malagasy people of Madagascar with a combined frequency of 34%.[13][14] O-M175 has been found in 28.1% of Solomon Islanders from Melanesia.[15] 12% of Uyghurs (Wells et al. 2001), 6.8% of Kalmyks[16] (17.1% of Khoshuud, 6.1% of Dörwöd, 3.3% of Torguud, 0% of Buzawa), 6.2% of Altaians (Kharkov et al. 2007), 4.1% of Uzbeks on average but Uzbeks from Bukhara 12.1%, Karakalpaks (Uzbekistan) 11.4%, Sinte (Uzbekistans) 6.7% (Wells et al. 2001) and 4.0% of Buryats.[17] In the Caucasus region it has been found in the Nogais 6%[18] but 5.3% in the Karan Nogais, it is also found in the Dargins of Dargwa speakers at 2.9%.[19] In the Iranic population, it is found in Iranian (Esfahan) at 6.3% (Wells et al. 2001), 8.9% of Tajiks in Afghanistan[20] 4.2% in the Pathans in Pakistan (Firasat 2007) but 1% in Afghanistan,[citation needed] 3.1% in Burusho (Firasat 2007).
Haplogroup O-M175 ranges in various moderate to high frequencies in the ethnic minorities of South Africa. The frequency of this haplogroup is 6.14% in the Cape colored population.[21] 18% in Cape Coloured Muslim, 38% in Cape Indian Muslims and 10% in other Cape Other Muslim.[21] It's found 11.5% in the Réunion Creole.[22] Haplogroup O-M175 had also been found in Latin America and Caribbean as a result of massive Chinese male migration from the 19th century. It was found in the Jamaicans at 3.8,[23] Cubans 1.5%[24]
Haplogroup O-M175 has been found in 88.7% of Asian American. 1.6% in Hispanic American, White Americans 0.5%, and 0.3% in African American.[25] Another study gives 0.5% African American.[26]
Among the sub-branches of haplogroup O-M175 are O-M119(O1a), O-M268(O1b), and O-M122(O2).
O-M175*
A broad survey of Y-chromosome variation among populations of central Eurasia found haplogroup O-M175(xM119,M95,M122) in 31% (14/45) of a sample of Koreans and in smaller percentages of Crimean Tatars (1/22 = 4.5%), Tajiks (1/16 = 6.25% Dushanbe, 1/40 = 2.5% Samarkand), Uyghurs (2/41 = 4.9%), Uzbeks (1/68 = 1.5% Surxondaryo, 1/70 = 1.4% Xorazm), and Kazakhs (1/54 = 1.9%) (Wells et al. 2001). However, nearly all of the purported Korean O-M175(xM119,M95,M122) Y-chromosomes may belong to Haplogroup O-M176,[Note 1] and later studies do not support the finding of O-M175* among similar population samples (Xue 2006, Kim 2011). The reported examples of O-M175(xM119,M95,M122) Y-chromosomes that have been found among these populations might therefore belong to Haplogroup O-M268*(xM95,M176) or Haplogroup O-M176 (O1b2).
A study published in 2013 found O-M175(xM119, M95, M176, M122) Y-DNA in 5.5% (1/18) Iranians from Teheran, 5.4% (2/37) Tajiks from Badakhshan Province of Afghanistan, and 1/97 Mongols from northwest Mongolia, while finding O-M176 only in 1/20 Mongols from northeast Mongolia.[27]
O-F265 (O1)
O1a-M119 and O1b-M268 share a common ancestor, O1-F265 (a.k.a. O-F75) approximately 33,181 (95% CI 24,461 to 36,879) YBP.[1][28] O1-F265, in turn, coalesces to a common ancestor with O2-M122 approximately 33,943 (95% CI 25,124 to 37,631) YBP.[1] Thus, O1-F265 should have existed as a single haplogroup parallel to O2-M122 for a duration of approximately 762 years (or anywhere from 0 to 13,170 years considering the 95% CIs and assuming that the phylogeny is correct) before breaking up into its two extant descendant haplogroups, O1-MSY2.2 and O1b-M268.
O-M119 (O1a)
O-M119 (which was known briefly as O-MSY2.2, until the SNP MSY2.2 was found to be unreliable) is found frequently in Austronesian-speaking people, with a moderate distribution in southern and eastern Chinese and Kra-dai peoples.
O-M268 (O1b)
- O-K18 Naxi[29]
- O-CTS4040
- O-MF56251 Observed sporadically in China (Guangxi,[29][30] Guangdong,[30] Sichuan,[30] Zhejiang,[30] Jiangsu,[30] Beijing[30]), Thailand (Phuan,[31] Yuan,[31] Central Thai[31]), Vietnam (Nùng,[31] Tày[31])
- O-Page59/CTS10887 Found among North Han Chinese (5%), East Han Chinese (4%), South Han Chinese (3%) [32]
- O-F4070
- O-MF106398 Observed sporadically in China (Guangdong, Henan, Hubei, Jiangxi, Sichuan, Zhejiang, Guangxi, Heilongjiang, Jiangsu, Shandong[30])
- O-F779/F993/F3135 China,[30] Vietnam (Lahu[33]), Qatar[34]
- O-MF107014 Observed sporadically in China (Jiangsu, Anhui, Heilongjiang[30])
- O-CTS5160/MF61620 China (Han,[29] mostly Guangdong or Fujian[30])
- O-F2064/F1759 China (Han from Fujian,[29] Shandong[29]), Singapore,[29] Vietnam (Sila,[33] Hanhi,[33] Kinh[33]), Korea[29]
- O-PH2797/CTS1127 China (especially Shandong, Jiangsu, Liaoning, Hebei, Anhui, Beijing, Henan, and Shanghai[30])
- O-Y148532 China (Shandong, Heilongjiang, Liaoning, Jilin, Jiangsu, Shanghai, Sichuan,[29] Beijing, Shaanxi[30]), Afghanistan (Hazara[29])
- O-Y239146/MF31164 Singapore,[29] Taiwan[29]
- O-Y47392/MF17288 China (Zhejiang[29])
- O-BY182144/Y157814 China (Shandong,[29][30] Gansu[30]), Taiwan[35]
- O-PH4822 China (Beijing,[29] Jiangsu[29])
- O-F417/M1654/CTS469 Japan (Tokyo[29])
- O-CTS9996/PF4341 Philippines[35]
- O-F4070
- O-PK4
- O-F838 Found in about 1.4% of Han Chinese[32] (and esp. in Hunan, Chongqing, Jiangxi, Sichuan, Guizhou[30])
- O-M95
- O-CTS350 China (Ningxia, Yunnan, Heilongjiang, Hunan, Shaanxi, Anhui, etc.[30])
- O-M1310
- O-Y172653/Y172877 Found in China (esp. Zhejiang, Fujian, Guangdong, Sichuan, Hunan, Jiangxi, Hubei, Chongqing[30]) and Japan[30]
- O-F1803/M1348 China (Zhejiang, Shandong, Beijing, Guangdong, Hubei, Sichuan, Jiangsu, Shanghai, etc.[30])
- O-ACT721/ACT1038 Found sporadically in China (Zhejiang,[30] Anhui,[30] Hunan,[30] Hainan,[29] Tianjin,[30] Beijing,[30] Liaoning,[29] Heilongjiang[30])
- O-F789/M1283 Found in China (Blang,[37] Palaung,[37] Wa,[37] Dai,[29][38] Yi,[29][38] Naxi), Vietnam, Cambodia, Singapore (Malay), Java, Borneo, Thailand, Laos, Myanmar, Bhutan, Bangladesh, India (Tripura, Ho, Konda Dora, Gond)
- O-M1283* Lao Isan[39]
- O-MF600645 Gansu (Hui,[29] Dongxiang[30]), Sichuan (Chengdu[30]), Hunan (Yiyang[30])
- O-M1368 Singapore[29]
- O-M1361
- O-MF611153 Found sporadically in China (Hunan, Hubei, Chongqing, Guangxi, Jiangxi[30])
- O-A22938 Vietnam (Kinh from Ho Chi Minh City[29]), China (Hong Kong,[29] Qinzhou,[30] Chongqing,[30] Lijiang[30])
- O-Y9322 China (Dai in Xishuangbanna,[29] Yunnan,[30] Chongqing,[30] Guangdong,[30] Sichuan,[30] etc.)
- O-Y9325
- O-Z39485 China (Dai, Yi)[29]
- O-Z39490
- O-Y9033/B426 Laos (Laotian[39]), Thailand (Blang,[39] Khmu,[39] Lawa,[39] Htin,[39] Padaung Karen,[39] Tai Dam,[39] Suay,[39] Khmer,[39] Mon,[39] Lao Isan,[39] Soa,[39] Shan,[39] Phutai,[39] Nyaw,[39] S'gaw Karen,[39] Thai,[39] Khon Mueang[39]), Vietnam (Mang from Mường Tè District,[33] Ede from Krông Buk District and Tuy An District,[33] Kinh from Hoàng Mai District, Gia Lâm District, and Yên Phong District,[33] Thái from Điện Biên Phủ,[33] Giarai from Ayun Pa[33])
- O-Y9325
- O-M1361
- O-F1252
- O-SK1630/F5504 China (esp. Sichuan and Guizhou, accounting for about 0.25% of the entire Chinese population[30])
- O-ACT5802
- O-MF92614
- O-F16061
- O-MF286118 Found in two Han Chinese from Guangdong[30]
- O-F19607
- O-ACT5802
- O-F2924
- O-CTS5854
- O-Z23810
- O-CTS7399
- O-Y85641 China (esp. Shandong, Liaoning, Jilin, Heilongjiang[30])
- O-Y14024/FGC19706 Japan (Tokyo[29])
- O-FGC19713/Y14026 Laos (Laotian in Vientiane and Luang Prabang[39]), Thailand (Tai Dam,[39] Tai Lue,[39] Nyah Kur,[39] Thai,[39] Eastern Lawa[39]), Vietnam (Thái from Bá Thước District and Tủa Chùa District,[33] Hà Nhì from Mường Tè District[33])
- O-FGC19707
- O-MF14427 China (esp. Jiangsu, Gansu, Henan, Shandong, Shanxi, Hebei, Shaanxi[30])
- O-FGC19716 China (Hong Kong,[29] Guangdong,[30] Hunan,[30] Chongqing[30])
- O-FGC19718 China (esp. Fujian, Jiangxi, Hubei, Jiangsu, Guangdong[30]), Philippines (Capiz[29])
- O-Z23849 China (Chongqing Han,[29] Xishuangbanna Dai,[29] Guangxi Zhuang,[29] Guangdong,[29] Shandong,[29] Tianjin[29])
- O-FGC19707
- O-CTS651/CTS10484 Thailand (Tai Khün,[39] Phuan,[39] Tai Lue,[39] Khon Mueang,[39] Eastern Lawa,[39] Lao Isan,[39] Thai[39]), Laos (Laotian in Vientiane[39]), Vietnam (Dao and Nùng from Hoàng Su Phì District,[33] Tày from Krông Pắk District, Hà Quảng District, and Đình Lập District[33])
- O-CTS7399
- O-Z23781 China (Henan[29])
- O-Z23810
- O-M111/M88 Found frequently among Vietnamese,[41][42][33] Tai peoples[43][44][42][33] (Bouyei,[43] Zhuang,[45][46][44] Nùng,[33] Tày,[33] Thái people in Vietnam,[33] Lao,[47] Northeastern Thai,[45] Northern Thai,[48][47] general population of Bangkok[42]), Lachi,[33] Lô Lô,[33] Hani-Akha,[43][42][33] Bunu,[49] She people,[41] Cambodians,[45] Kuy,[37][49] Bru,[49] and Htin,[37] with a moderate distribution among Qiang,[43] Bai,[50] Yi,[44] Bamar,[51] Jingpo,[51] Lahu,[33] Tujia,[45] Han Chinese,[45][43][41][42] Miao,[41][49] Pathen,[33] Yao,[43][41][49][33] Hlai,[45][43] Taiwanese aborigines[13][41][42] (especially Bunun[48][42]), the Philippines,[41][42] Malaysia[41] (Kota Kinabalu[13]), Kalimantan (Banjarmasin[13]), Java,[42] Chamic-speaking peoples (Cham from Bình Thuận,[47] Ede,[33] Jarai[33]), and Kiribati[48]
- O-M111/M88* Northern Thailand (Htin, Lawa), Cambodia (Jarai, Brao, Kachac, Khmer, Lao, Lun), Yunnan (De'ang)[37]
- O-F2524
- O-F2524* Jiangsu[29]
- O-F2346
- O-F2890 Thailand (Khon Mueang,[39] Phuan,[39] Shan,[39] Htin,[39] Tai Dam,[39] Thai,[39] Lawa,[39] Lao Isan,[39] Mon[39]), Vietnam (Kinh from Gia Lâm District,[33] Tày from Lục Yên District[33])
- O-F2890* Ho Chi Minh City[29]
- O-Z24048
- O-F2758 Vietnam (Kinh,[33] Lahu,[33] Dao, Pathen,[33] Tày,[33] Thái,[33] Ede,[33] Giarai[33]), Cambodia (Kuy, Tampuan, Khmer), Thailand (Phutai,[39] Bru,[39] Tai Khün,[39] Phuan,[39] Tai Dam,[39] Shan,[39] Khon Mueang,[39] Mon,[39] Lao Isan,[39] Tai Lue,[39] Htin,[37][39] Lawa,[37] Khmu,[39] Kaleun,[39] Nyaw,[39] Suay,[39] Thai[39]), Laos (Laotian in Luang Prabang[39]), Yunnan (Bulang, De'ang)[37]
- O-F2758* China (Miao,[29] Hunan[29])
- O-Z24083
- O-Z24083* Ho Chi Minh City (Kinh)
- O-Z24089
- O-SK1627/Z24091 Vietnam (Lô Lô from Mèo Vạc District,[33] La Chí and Nùng from Hoàng Su Phì District,[33] Hà Nhì from Mường Tè District,[33] Tày from Chợ Đồn District and Đăk Mil District,[33] Ede from Ea Kar District,[33] Kinh from Nghĩa Hưng District[33]), Thailand (Soa,[39] Saek,[39] Phutai,[39] Suay,[39] Tai Dam,[39] S'gaw Karen,[39] Nyah Kur,[39] Khmer,[39] Lawa,[39] Lao Isan,[39] Mon,[39] Thai[39]), Laos (Laotian in Vientiane[39])
- O-F923
- O-F2890 Thailand (Khon Mueang,[39] Phuan,[39] Shan,[39] Htin,[39] Tai Dam,[39] Thai,[39] Lawa,[39] Lao Isan,[39] Mon[39]), Vietnam (Kinh from Gia Lâm District,[33] Tày from Lục Yên District[33])
- O-CTS5854
- O-SK1630/F5504 China (esp. Sichuan and Guizhou, accounting for about 0.25% of the entire Chinese population[30])
- O-CTS4040
- O-M176
- O-K4: Found frequently among Koreans and with a moderate distribution among Japanese, Ryukyuans, Daurs, Evenks, Hezhe, Manchus, and Sibe. Also found sporadically (<1%) among Han Chinese, Hui, Micronesians, Mongols, Thais, Uyghurs, Vietnamese, etc.
- O-47z: Found frequently among Japanese and Ryukyuans and with a moderate distribution among Koreans. Found sporadically (<1%) among Manchus, Mongols, Han Chinese, Hui, Tujia, Vietnamese, etc.
O-M122 (O2)
Found frequently among populations of East Asia, Southeast Asia, and culturally Austronesian regions of Oceania, with a moderate distribution in Central Asia (Shi 2005).
- O-M122
- O-CTS1754 East & Southeast Asia
- O-M324
- O-L465
- O-CTS727
- O-F915
- O-CTS3709
- O-JST002611/CTS2483
- O-CTS2483* China,[35] Japan,[35] Philippines[35]
- O-CTS10573 Beijing,[52] Sichuan,[52] Henan,[52] Jiangsu[52]
- O-F18
- O-CTS498 China,[29] Japan (Tokyo)[29]
- O-F449 Azerbaijan[35]
- O-F117
- O-F117* Fujian[29]
- O-F11
- O-F11* Gansu,[29] Japanese[29]
- O-F930 Beijing,[29] Armenia,[29] Inner Mongolia,[52] Hebei,[52] Shaanxi,[52] Shandong,[52] Zhejiang,[52] Hubei[52]
- O-F2685 Beijing, Shanghai, Fujian, Guangdong[52]
- O-BY169374
- O-F539 Beijing, Shanghai, Jiangsu, Zhejiang, Jiangxi, Guangdong, Yunnan[52]
- O-CTS12877
- O-Y29837
- O-BY36917 Japan[35]
- O-F4062 Beijing, Shanghai, Guangdong, Jiangsu, Shandong, Shaanxi, Chongqing, Heilongjiang, Liaoning, Henan, Hubei, Hunan, Zhejiang[52]
- O-Y15976 China, Japan,[35] Korea, Pakistan, Vietnam
- O-FGC54474
- O-F971 Beijing, Shanghai, Hubei, Guangdong[52]
- O-F632
- O-F632* Beijing[29]
- O-F16340 Zhejiang[29]
- O-F133 China, Bulgaria[35]
- O-CTS727
- O-P201
- O-M188
- O-M188* Korea[35]
- O-CTS800
- O-CTS445
- O-CTS201 Korea[35]
- O-M159 China (about 0.79% of the national male population[53]), Taiwan, Cambodia, Malaysia, Singapore[35]
- O-FTA21663/O-MF22947 China (Heilongjiang,[29][30] Inner Mongolia,[29][30] Zhejiang,[30] Shanghai,[30] Henan,[30] Hebei,[30] etc.; accounts for about 0.06% of the male population in China at present[54]), Saudi Arabia (al-Qaṣīm[29])
- O-CTS3994
- O-MF18110/FGC50590 China (esp. Guangdong, Zhejiang, Hunan, Shandong, and Guangxi[30])
- O-MF109844
- O-FGC50661 China (esp. Jiangsu and Hunan[30])
- O-MF56709
- O-FGC50643/MF15475 China (Shandong,[30] Hebei,[30] Hubei,[30] Shanxi,[29][30] Anhui,[30] Jiangsu,[30] etc.)
- O-MF56474 China (Jiangsu, Anhui, Jilin, Shandong, etc.[30])
- O-FGC50649
- O-Y169670/O-MF14256 China (esp. Jiangsu,[29][30] Shandong,[30] Zhejiang,[30] and Shanghai[30])
- O-MF50824
- O-MF14135/O-Z12303 China (currently accounts for about 0.44% of the total male population[56])
- O-MF238642
- O-MF37094 China (Zhejiang,[30] Jiangsu[30])
- O-Y169696/O-MF15693 China (Jiangsu,[29][30] Fufeng County,[30] Beijing,[30] Tangshan,[30] Feidong County,[30] Chifeng,[30] Xi County,[30] Min County,[30] Laizhou,[30] Rushan,[30] Harbin,[30] Yanji,[30] Dancheng County[30])
- O-MF18577/O-MF18626 China (currently accounts for about 0.23% of all males in China, especially in Jiangsu [1.08%], Shanghai [0.69%], Ningxia [0.39%], Shandong [0.38%], Anhui [0.33%], Heilongjiang [0.32%], Zhejiang [0.31%], and Jilin [0.26%][57]), Kazakhstan,[58] Thailand[58]
- O-MF238642
- O-FGC50558 Japan,[35] Korea[35]
- O-Y169670/O-MF14256 China (esp. Jiangsu,[29][30] Shandong,[30] Zhejiang,[30] and Shanghai[30])
- O-M159 China (about 0.79% of the national male population[53]), Taiwan, Cambodia, Malaysia, Singapore[35]
- O-M7 Found frequently among human remains associated with the Neolithic Daxi culture[59] and modern Hmong–Mien, Katuic, and Bahnaric peoples,[49] with a moderate distribution among Han Chinese (Xue 2006), Buyei (Xue 2006), Bai (Wen 2004), Mosuo (Wen 2004), Tibetans (Wen 2004), Qiang (Xue 2006), Oroqen (Xue 2006), Tujia (Su 2000), Thai (Su 2000), Orang Asli (Su 2000), western Indonesians (Su 2000 and Kayser 2008), Malaysians (Kayser 2008), Vietnamese (Kayser 2008), and Atayal (Su 2000).
- O-MF106687 China (Jinghu District,[30] etc.)
- O-Z25245
- O-MF9858/O-Z6157 China (approximately 0.08% of all males in present-day China[60]), Thailand (Central Thai in Central Thailand[39][31])
- O-Y26422
- O-F1276
- O-F1863
- O-MF107102 China (Tongchuan District[30])
- O-MF56735 China (Haiyan County, Suzhou, Wuxi, Shanghai, etc.[30])
- O-MF36531 China (Han in Yanping District[30])
- O-F1134
- O-MF35799/O-Y94171 Thailand (Mon in Central Thailand[39][31]), China (observed sporadically in Pingyang County, Yunan County, Siming District, Longyao County, Shanwei, etc.[30])
- O-F1262/O-Y173492 China[29][58] (accounts for about 0.15% of the male population in China at present and is relatively concentrated in Zhejiang, Taiwan, Anhui, Jiangxi, etc.;[61] also observed in individuals from Zhenjiang,[31] Hejian,[31] and Langfang[31])
- O-FT303223/O-MF106843/O-F15314/O-F20756 China[58] (Changsha,[30] Chancheng District,[30] Wanzhou District,[30] Chaoyang District,[30] Dongying,[30] Chifeng,[30] Liaoyuan,[30] Harbin,[30] Han in Zhengzhou,[31] Dai in Xishuangbanna[29]), Thailand[58] (Khon Mueang in Northern Thailand,[39][31] Black Tai in Loei Province[39][31])
- O-Z25288/O-Z25293 Vietnam[58] (Kinh in Ho Chi Minh City,[29][31] Hanoi,[31] Nam Dinh,[31] and Lao Cai,[31] Giarai in Gia Lai,[31] Tày in Thai Nguyen[31])
- O-CTS6489
- O-MF106428/O-Y94472/O-FTB23660 Thailand[58] (Phayao,[29] Phutai, Lao Isan, Tai Lue, Phuan, Shan, Khon Mueang/Tai Yuan, Khmer, Mon[31]), Vietnam[58] (Tày in Lào Cai[31]), China (Dai in Xishuangbanna,[29] Achang in Yunnan;[31] accounts for about 0.05% of all males in China at present, mainly distributed in Guangxi and Guangdong[62])
- O-F1275 Guangxi (Dushan 4-1 ca. 7024 - 6643 BCE[58])
- O-MF15199/O-FTA25885
- O-F20472
- O-FTB23785 Thailand,[58] Vietnam[58]
- O-F17410/O-F18833/O-MF122643/O-BY177553 Thailand (Lao Isan in Northeast Thailand[39][31])
- O-MF106415/O-MF111486/O-BY122399 Thailand[58] (Shan in Mae Hong Son Province[63][31]), China (observed sporadically in individuals from Hubei, Hunan, Chongqing, Sichuan, Guangxi, Jiangxi, Zhejiang, Jiangsu, Anhui, Gansu, Shaanxi, Shanxi, Henan, and Shandong[30])
- O-Y127482/O-F15988 Thailand (Nyahkur and Lao Isan in Northeast Thailand[39][31])
- O-MF6534/O-MF58872/O-BY27925/O-Y23477 Thailand[58] (Central Thai in Central Thailand, Phuan in Central Thailand, Khon Mueang in North Thailand,[39][31] White Hmong in Chiang Rai Province[63][31]), Singapore,[29][31] China (Guangdong, Guangxi, Hunan, Sichuan, Jiangxi, Zhejiang, Shaanxi, Henan, Shandong[31][30])
- O-CTS6579
- O-CTS123/O-F22573/O-MF48275 China (Hunan Han;[29] accounts for about 0.13% of the male population in China at present, mainly distributed in Jiangxi, Hunan and other south-central provinces and cities[64])
- O-F14832/O-F15788/O-Y208219 China[58] (accounts for about 0.22% of the male population in China at present, mainly distributed in the northern region[65]), Thailand[58] (Mon in Western Thailand,[39][31] Tai Lue in Northern Thailand[39][31])
- O-F20472
- O-Z25411
- O-ACT1126/O-Y140772/O-F1289 China (relatively concentrated in northern China at present, accounting for about 0.24% of the national male population;[66] also found in Fujian[29]), Thailand[58] (Lisu[31])
- O-Z25398
- O-F22005/O-Z25400 Thailand[58] (Black Hmong in North Thailand[29][63]), Vietnam[58] (Kinh in Ho Chi Minh City[29]), China (currently distributed mainly in Guangxi, Sichuan, Guangdong and other places, accounting for about 0.10% of the national male population[67])
- O-F1100/O-Y37861 Hunan[29]
- O-MF17697 Laos,[58] Thailand,[58] China (Jiangsu, Hunan, Jiangxi, Guangxi, Guangdong, Guizhou, Yunnan, Fujian, Sichuan, Hong Kong, Chongqing, Henan, Liaoning[30])
- O-F1234/O-Y37855
- O-Y185160/O-MF36985 Hebei,[29] Beijing,[29] Sichuan, Shaanxi, Guangxi, Zhejiang, Shandong, Ningxia, Inner Mongolia, Hubei, Jiangxi (currently accounts for approximately 0.13% of the Chinese male population[68])
- O-FGC71370
- O-MF193618 Sichuan, Zhejiang, Shandong, Anhui, Hunan, Hubei, Fujian (currently accounts for about 0.08% of the male population in China, mainly distributed in Guangdong, Hunan, Anhui and other provinces and cities[69]), Philippines[58]
- O-F14904/N5 Ningxia,[29] Hmong (Northern Thailand[63]), She,[29] Iu Mien (Phayao Province[63]), Quebec[29]. Huang et al. (2022) found that this is the most common Y-chromosome haplogroup among many Hmongic-speaking ethnic groups (including Guangxi Miao, Hunan Miao, Hunan Pa-hng, and Thailand Hmong), with a frequency of 47.1% among the Guangxi Miao.[70]
- O-MF15199/O-FTA25885
- O-F1863
- O-CTS201 Korea[35]
- O-P164
- O-F996/F3237
- O-A16433 Heilongjiang[29]
- O-MF56976 Anhui[29]
- O-Y125645
- O-F871
- O-A16433 Heilongjiang[29]
- O-M134: Found frequently among speakers of Sino-Tibetan languages, among members of the Kazakh Naiman tribe with a moderate distribution throughout East Asia and Southeast Asia.[citation needed]
- O-Y20/PAGES00125 Poland[35]
- O-F1725
- O-Y12/F314
- O-Y12* Beijing (Han)[29]
- O-CTS2643/CTS11192
- O-CTS53
- O-F876
- O-F275
- O-F634
- O-CTS3776/F2887
- O-M117/PAGE23
- O-MF1380/CTS4960 China, Korea, Japan,[35] Indonesia[35]
- O-M133/M1706 Shandong[29]
- O-M1706* Japan (Tokyo)[29]
- O-YP4864
- O-CTS7634
- O-M1726
- O-A9459
- O-F6800
- O-F14249
- O-F438 Japan (Tokyo)[29]
- O-CTS1642
- O-Y20/PAGES00125 Poland[35]
- O-F996/F3237
- O-M188
- O-L465
O-M324 (O2a)
O-F742 (O2b)
Language families and genes
Haplogroup O is associated with populations which speak Austric languages. The following is a phylogenetic tree of language families and their corresponding SNP markers, or haplogroups, sourced mainly from Edmondson 2007 and Shi 2005. This has been called the "Father Tongue Hypothesis" by George van Driem (van Driem 2011). It does not appear to account for O-M176, which is found among Japanese, Korean, and Manchurian males.
| (M175) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Phylogenetics
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being, above all, timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O-M175 | 26 | VII | 1U | 28 | Eu16 | H9 | I | O* | O | O | O | O | O | O | O | O | O | O |
| O-M119 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1* | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a | O1a |
| O-M101 | 26 | VII | 1U | 32 | Eu16 | H9 | H | O1a | O1a1 | O1a1a | O1a1a | O1a1 | O1a1 | O1a1a | O1a1a | O1a1a | O1a1a | O1a1a |
| O-M50 | 26 | VII | 1U | 32 | Eu16 | H10 | H | O1b | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 | O1a2 |
| O-P31 | 26 | VII | 1U | 33 | Eu16 | H5 | I | O2* | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 | O2 |
| O-M95 | 26 | VII | 1U | 34 | Eu16 | H11 | G | O2a* | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a | O2a1 | O2a1 |
| O-M88 | 26 | VII | 1U | 34 | Eu16 | H12 | G | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1 | O2a1a | O2a1a |
| O-SRY465 | 20 | VII | 1U | 35 | Eu16 | H5 | I | O2b* | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b | O2b |
| O-47z | 5 | VII | 1U | 26 | Eu16 | H5 | I | O2b1 | O2b1a | O2b1 | O2b1 | O2b1a | O2b1a | O2b1 | O2b1 | O2b1 | O2b1 | O2b1 |
| O-M122 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3* | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 | O3 |
| O-M121 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3a | O3a | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1 | O3a1a | O3a1a |
| O-M164 | 26 | VII | 1U | 29 | Eu16 | H6 | L | O3b | O3b | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a2 | O3a1b | O3a1b |
| O-M159 | 13 | VII | 1U | 31 | Eu16 | H6 | L | O3c | O3c | O3a3a | O3a3a | O3a3 | O3a3 | O3a3a | O3a3a | O3a3a | O3a3a | O3a3a |
| O-M7 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d* | O3c | O3a3b | O3a3b | O3a4 | O3a4 | O3a3b | O3a3b | O3a3b | O3a2b | O3a2b |
| O-M113 | 26 | VII | 1U | 29 | Eu16 | H7 | L | O3d1 | O3c1 | O3a3b1 | O3a3b1 | - | O3a4a | O3a3b1 | O3a3b1 | O3a3b1 | O3a2b1 | O3a2b1 |
| O-M134 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e* | O3d | O3a3c | O3a3c | O3a5 | O3a5 | O3a3c | O3a3c | O3a3c | O3a2c1 | O3a2c1 |
| O-M117 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1* | O3d1 | O3a3c1 | O3a3c1 | O3a5a | O3a5a | O3a3c1 | O3a3c1 | O3a3c1 | O3a2c1a | O3a2c1a |
| O-M162 | 26 | VII | 1U | 30 | Eu16 | H8 | L | O3e1a | O3d1a | O3a3c1a | O3a3c1a | O3a5a1 | O3a5a1 | O3a3c1a | O3a3c1a | O3a3c1a | O3a2c1a1 | O3a2c1a1 |
Original Research Publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Phylogenetic trees
ISOGG 2017 tree (ver. 12.244).[74]
- O (M175)
- O1 (F265/M1354, CTS2866, F75/M1297, F429/M1415, F465/M1422)
- O1a (M119)
- O1a1 (B384/Z23193)
- O1a1a (M307.1/P203.1)
- O1a1a1 (F446)
- O1a1a1a (F140)
- O1a1a1a1 (F78)
- O1a1a1a1a (F81)
- O1a1a1a1a1 (CTS2458)
- O1a1a1a1a1a (F533)
- O1a1a1a1a1a1 (F492)
- O1a1a1a1a1a1a (F656)
- O1a1a1a1a1a1a1 (A12440)
- O1a1a1a1a1a1a1a (A12439)
- O1a1a1a1a1a1a2 (A14788)
- O1a1a1a1a1a1a3 (F65)
- O1a1a1a1a1a1a4 (MF1068)
- O1a1a1a1a1a1a5 (Z23482)
- O1a1a1a1a1a1a1 (A12440)
- O1a1a1a1a1a1b (FGC66168)
- O1a1a1a1a1a1b1 (CTS11553)
- O1a1a1a1a1a1c (Y31266)
- O1a1a1a1a1a1c1 (Y31261)
- O1a1a1a1a1a1d (A12441)
- O1a1a1a1a1a1e (MF1071)
- O1a1a1a1a1a1e1 (MF1074)
- O1a1a1a1a1a1a (F656)
- O1a1a1a1a1a2 (CTS4585)
- O1a1a1a1a1a1 (F492)
- O1a1a1a1a1a (F533)
- O1a1a1a1a2 (MF1075)
- O1a1a1a1a1 (CTS2458)
- O1a1a1a1a (F81)
- O1a1a1a2 (YP4610/Z39229)
- O1a1a1a2a (AM00330/AMM480/B386)
- O1a1a1a2a1 (AM00333/AMM483/B387)
- O1a1a1a2a1a (B388)
- O1a1a1a2a1 (AM00333/AMM483/B387)
- O1a1a1a2b (SK1555)
- O1a1a1a2a (AM00330/AMM480/B386)
- O1a1a1a1 (F78)
- O1a1a1b (SK1568/Z23420)
- O1a1a1b1 (M101)
- O1a1a1b2 (Z23392)
- O1a1a1b2a (Z23442)
- O1a1a1b2a1 (SK1571)
- O1a1a1b2a (Z23442)
- O1a1a1a (F140)
- O1a1a2 (CTS52)
- O1a1a2a (CTS701)
- O1a1a2a1 (K644/Z23266)
- O1a1a2a (CTS701)
- O1a1a1 (F446)
- O1a1b (CTS5726)
- O1a1a (M307.1/P203.1)
- O1a2 (M110)
- O1a2a (F3288)
- O1a2a1 (B392)
- O1a2a1a (B393)
- O1a2a1 (B392)
- O1a2a (F3288)
- O1a3 (Page109)
- O1a1 (B384/Z23193)
- O1b (M268)
- O1b1 (F2320)
- O1b1a (M1470)
- O1b1a1 (PK4)
- O1b1a1a (M95)
- O1b1a1a1 (F1803/M1348)
- O1b1a1a1a (F1252)
- O1b1a1a1a1 (F2924)
- O1b1a1a1a1a (M111)
- O1b1a1a1a1a1 (F2758)
- O1b1a1a1a1a1a (Z24083)
- O1b1a1a1a1a1a1 (Z24089)
- O1b1a1a1a1a1a1a (F923)
- O1b1a1a1a1a1a1a1 (CTS2022)
- O1b1a1a1a1a1a1a1a (F1399)
- O1b1a1a1a1a1a1a1a1 (F2415)
- O1b1a1a1a1a1a1a1a (F1399)
- O1b1a1a1a1a1a1a2 (Z24131)
- O1b1a1a1a1a1a1a3 (Z24100)
- O1b1a1a1a1a1a1a1 (CTS2022)
- O1b1a1a1a1a1a1b (SK1627/Z24091)
- O1b1a1a1a1a1a1b1 (Z39410)
- O1b1a1a1a1a1a1a (F923)
- O1b1a1a1a1a1a2 (Z24088)
- O1b1a1a1a1a1a1 (Z24089)
- O1b1a1a1a1a1a (Z24083)
- O1b1a1a1a1a2 (F2890)
- O1b1a1a1a1a2a (Z24048)
- O1b1a1a1a1a2a1 (Z24050)
- O1b1a1a1a1a2b (Z24014)
- O1b1a1a1a1a2a (Z24048)
- O1b1a1a1a1a1 (F2758)
- O1b1a1a1a1b (CTS5854)
- O1b1a1a1a1b1 (Z23810)
- O1b1a1a1a1b1a (CTS7399)
- O1b1a1a1a1b1a1 (FGC19713/Y14026)
- O1b1a1a1a1b1a1a (Z23849)
- O1b1a1a1a1b1a1a1 (FGC61038)
- O1b1a1a1a1b1a1a (Z23849)
- O1b1a1a1a1b1a1 (FGC19713/Y14026)
- O1b1a1a1a1b1b (CTS651)
- O1b1a1a1a1b1b1 (CTS9884)
- O1b1a1a1a1b1a (CTS7399)
- O1b1a1a1a1b2 (F4229)
- O1b1a1a1a1b2a (F809)
- O1b1a1a1a1b2a1 (F2517)
- O1b1a1a1a1b2a (F809)
- O1b1a1a1a1b1 (Z23810)
- O1b1a1a1a1a (M111)
- O1b1a1a1a2 (SK1630)
- O1b1a1a1a2a (SK1636)
- O1b1a1a1a1 (F2924)
- O1b1a1a1b (F789/M1283)
- O1b1a1a1b1 (FGC29900/Y9322/Z23667)
- O1b1a1a1b1a (B426/FGC29896/Y9033/Z23671)
- O1b1a1a1b1a1 (FGC29907/YP3930)
- O1b1a1a1b1a2 (B427/Z23680)
- O1b1a1a1b1b (Z39485)
- O1b1a1a1b1c (B418)
- O1b1a1a1b1a (B426/FGC29896/Y9033/Z23671)
- O1b1a1a1b2 (SK1646)
- O1b1a1a1b1 (FGC29900/Y9322/Z23667)
- O1b1a1a1a (F1252)
- O1b1a1a2 (CTS350)
- O1b1a1a3 (Page103)
- O1b1a1a1 (F1803/M1348)
- O1b1a1b (F838)
- O1b1a1b1 (F1199)
- O1b1a1a (M95)
- O1b1a2 (Page59)
- O1b1a2a (F993)
- O1b1a2a1 (F1759)
- O1b1a2a1a (CTS1127)
- O1b1a2a1 (F1759)
- O1b1a2b (F417/M1654)
- O1b1a2b1 (F840)
- O1b1a2b1a (F1127)
- O1b1a2b2 (CTS1451)
- O1b1a2b1 (F840)
- O1b1a2c (CTS9996)
- O1b1a2a (F993)
- O1b1a1 (PK4)
- O1b1a (M1470)
- O1b2 (P49, M176)
- O1b2a (F1942/Page92)
- O1b2a1 (CTS9259)
- O1b2a1a (F1204)
- O1b2a1a1 (CTS713)
- O1b2a1a1a (CTS1875)
- O1b2a1a1a1 (CTS10682)
- O1b2a1a1b (Z24598)
- O1b2a1a1c (CTS203)
- O1b2a1a1a (CTS1875)
- O1b2a1a2 (F2868)
- O1b2a1a2a (L682)
- O1b2a1a2a1 (CTS723)
- O1b2a1a2a1a (CTS7620)
- O1b2a1a2a1b (A12446)
- O1b2a1a2a1b1 (PH40)
- O1b2a1a2a1 (CTS723)
- O1b2a1a2b (F940)
- O1b2a1a2a (L682)
- O1b2a1a3 (CTS10687)
- O1b2a1a3a (CTS1215)
- O1b2a1a1 (CTS713)
- O1b2a1b (CTS562)
- O1b2a1a (F1204)
- O1b2a2 (Page90)
- O1b2a1 (CTS9259)
- O1b2a (F1942/Page92)
- O1b1 (F2320)
- O1a (M119)
- O2 (M122)
- O2a (M324)
- O2a1 (L127.1)
- O2a1a (F1876/Page127)
- O2a1a1 (F2159)
- O2a1a1a (F1867/Page124)
- O2a1a1a1 (F852)
- O2a1a1a1a (F2266)
- O2a1a1a1a1 (L599)
- O2a1a1a1a1a (Z43961)
- O2a1a1a1a1a1 (Z43963)
- O2a1a1a1a1a (Z43961)
- O2a1a1a1a1 (L599)
- O2a1a1a1b (F854)
- O2a1a1a1b1 (Z43966)
- O2a1a1a1c (Page130)
- O2a1a1a1a (F2266)
- O2a1a1a1 (F852)
- O2a1a1b (F915)
- O2a1a1b1 (F1478)
- O2a1a1b1a (PF5390)
- O2a1a1b1a1 (CTS1936)
- O2a1a1b1a1a (Z43975)
- O2a1a1b1a2 (FGC33994)
- O2a1a1b1a (PF5390)
- O2a1a1b1 (F1478)
- O2a1a1a (F1867/Page124)
- O2a1a1 (F2159)
- O2a1b (M164)
- O2a1c (IMS-JST002611)
- O2a1c1 (F18)
- O2a1c1a (F117)
- O2a1c1a1 (F13)
- O2a1c1a1a (F11)
- O2a1c1a1a1 (F632)
- O2a1c1a1a1a (F110/M11115)
- O2a1c1a1a1a1 (F17)
- O2a1c1a1a1a1a (F377)
- O2a1c1a1a1a1a1 (F1095)
- O2a1c1a1a1a1a1a (F856)
- O2a1c1a1a1a1a1a1 (F1418)
- O2a1c1a1a1a1a1a2 (Z25097)
- O2a1c1a1a1a1a1a (F856)
- O2a1c1a1a1a1a2 (CTS7501)
- O2a1c1a1a1a1a1 (F1095)
- O2a1c1a1a1a1b (F793)
- O2a1c1a1a1a1a (F377)
- O2a1c1a1a1a2 (Y20951)
- O2a1c1a1a1a2a (Y20932)
- O2a1c1a1a1a1 (F17)
- O2a1c1a1a1a (F110/M11115)
- O2a1c1a1a2 (F38)
- O2a1c1a1a3 (F12)
- O2a1c1a1a4 (F930)
- O2a1c1a1a4a (F2685)
- O2a1c1a1a5 (F1365/M5420/PF1558)
- O2a1c1a1a5a (Y15976)
- O2a1c1a1a5a1 (Y16154)
- O2a1c1a1a5a1a (Y26383)
- O2a1c1a1a5a1a1 (SK1686)
- O2a1c1a1a5a1a (Y26383)
- O2a1c1a1a5a1 (Y16154)
- O2a1c1a1a5b (FGC54486)
- O2a1c1a1a5b1 (FGC54507)
- O2a1c1a1a5a (Y15976)
- O2a1c1a1a6 (CTS12877)
- O2a1c1a1a6a (F2527)
- O2a1c1a1a6a1 (CTS5409)
- O2a1c1a1a6a2 (F2941)
- O2a1c1a1a6a (F2527)
- O2a1c1a1a7 (F723)
- O2a1c1a1a8 (CTS2107)
- O2a1c1a1a9 (SK1691)
- O2a1c1a1a1 (F632)
- O2a1c1a1b (PH203)
- O2a1c1a1a (F11)
- O2a1c1a1 (F13)
- O2a1c1b (F449)
- O2a1c1b1 (F238)
- O2a1c1b1a (F134)
- O2a1c1b1a1 (F1273)
- O2a1c1b1a2 (F724)
- O2a1c1b1a (F134)
- O2a1c1b2 (F1266)
- O2a1c1b1 (F238)
- O2a1c1c (CTS498)
- O2a1c1a (F117)
- O2a1c2 (FGC3750/SK1673)
- O2a1c1 (F18)
- O2a1a (F1876/Page127)
- O2a2 (IMS-JST021354/P201)
- O2a2a (M188)
- O2a2a1 (F2588)
- O2a2a1a (CTS445)
- O2a2a1a1 (CTS201)
- O2a2a1a1a (M159/Page96)
- O2a2a1a2 (M7)
- O2a2a1a2a (F1276)
- O2a2a1a2a1 (CTS6489)
- O2a2a1a2a1a (F1275)
- O2a2a1a2a1a1 (M113)
- O2a2a1a2a1a2 (N5)
- O2a2a1a2a1a3 (Z25400)
- O2a2a1a2a1a (F1275)
- O2a2a1a2a2 (F1863)
- O2a2a1a2a2a (F1134)
- O2a2a1a2a2a1 (F1262)
- O2a2a1a2a2a (F1134)
- O2a2a1a2a1 (CTS6489)
- O2a2a1a2b (Y26403)
- O2a2a1a2a (F1276)
- O2a2a1a1 (CTS201)
- O2a2a1b (F1837)
- O2a2a1a (CTS445)
- O2a2a2 (F879)
- O2a2a2a (F1226)
- O2a2a2a1 (F2859)
- O2a2a2a (F1226)
- O2a2a1 (F2588)
- O2a2b (P164)
- O2a2b1 (M134)
- O2a2b1a (F450/M1667)
- O2a2b1a1 (M117/Page23)
- O2a2b1a1a (M133)
- O2a2b1a1a1 (F438)
- O2a2b1a1a1a (Y17728)
- O2a2b1a1a1a1 (F155)
- O2a2b1a1a1a1a (F813/M6539)
- O2a2b1a1a1a1a1 (Y20928)
- O2a2b1a1a1a1a (F813/M6539)
- O2a2b1a1a1a2 (F1754)
- O2a2b1a1a1a2a (F2137)
- O2a2b1a1a1a2a1 (F1442)
- O2a2b1a1a1a2a1a (F1123)
- O2a2b1a1a1a2a1a1 (F1369)
- O2a2b1a1a1a2a1a (F1123)
- O2a2b1a1a1a2a2 (A16636)
- O2a2b1a1a1a2a1 (F1442)
- O2a2b1a1a1a2a (F2137)
- O2a2b1a1a1a3 (Z25907)
- O2a2b1a1a1a1 (F155)
- O2a2b1a1a1a (Y17728)
- O2a2b1a1a2 (FGC23469/Z25852)
- O2a2b1a1a2a (F310)
- O2a2b1a1a2a1 (F402)
- O2a2b1a1a2a1a (F1531)
- O2a2b1a1a2a1 (F402)
- O2a2b1a1a2a (F310)
- O2a2b1a1a3 (CTS7634)
- O2a2b1a1a3a (F317)
- O2a2b1a1a3a1 (F3039)
- O2a2b1a1a3a2 (Y29861)
- O2a2b1a1a3b (CTS5488)
- O2a2b1a1a3a (F317)
- O2a2b1a1a4 (Z25853)
- O2a2b1a1a4a (CTS5492)
- O2a2b1a1a4a1 (CTS6987)
- O2a2b1a1a4a1a (Z42620)
- O2a2b1a1a4a2 ( F20963)
- O2a2b1a1a4a1 (CTS6987)
- O2a2b1a1a4a (CTS5492)
- O2a2b1a1a5 (CTS10738/M1707)
- O2a2b1a1a5a (CTS9678)
- O2a2b1a1a5a1 (Z39663)
- O2a2b1a1a5a2 (M1513)
- O2a2b1a1a5b (A9457)
- O2a2b1a1a5b1 (F17158)
- O2a2b1a1a5a (CTS9678)
- O2a2b1a1a6 (CTS4658)
- O2a2b1a1a6a (CTS5308)
- O2a2b1a1a6b (Z25928)
- O2a2b1a1a6b1 (SK1730/Z25982)
- O2a2b1a1a6b1a (Z26030)
- O2a2b1a1a6b1b (Z26010)
- O2a2b1a1a6b2 (A9462)
- O2a2b1a1a6b3 (B456)
- O2a2b1a1a6b1 (SK1730/Z25982)
- O2a2b1a1a7 (YP4864)
- O2a2b1a1a7a (Z44068)
- O2a2b1a1a7a1 (F5525/SK1748)
- O2a2b1a1a7b (Z44071)
- O2a2b1a1a7a (Z44068)
- O2a2b1a1a8 (Z44091)
- O2a2b1a1a8a (Z44092)
- O2a2b1a1a1 (F438)
- O2a2b1a1b (CTS4960)
- O2a2b1a1a (M133)
- O2a2b1a2 (F114)
- O2a2b1a2a (F79)
- O2a2b1a2a1 (F46/Y15)
- O2a2b1a2a1a (FGC16847/Z26091)
- O2a2b1a2a1a1 (F48)
- O2a2b1a2a1a1a (F152)
- O2a2b1a2a1a1a1 (F2505)
- O2a2b1a2a1a1b (CTS3149)
- O2a2b1a2a1a1a (F152)
- O2a2b1a2a1a2 (F242)
- O2a2b1a2a1a2a (CTS4266)
- O2a2b1a2a1a2a1 (Z26108)
- O2a2b1a2a1a2a1a (F2173)
- O2a2b1a2a1a2a1 (Z26108)
- O2a2b1a2a1a2a (CTS4266)
- O2a2b1a2a1a3 (F2887)
- O2a2b1a2a1a3a (F3607)
- O2a2b1a2a1a3a1 (F3525)
- O2a2b1a2a1a3b (CTS3763)
- O2a2b1a2a1a3b1 (A9472)
- O2a2b1a2a1a3b2 (FGC16863/Y7110)
- O2a2b1a2a1a3b2a (L1360)
- O2a2b1a2a1a3b2a1 (FGC16889)
- O2a2b1a2a1a3b2b (SK1768/Y7112/Z26257)
- O2a2b1a2a1a3b2b1 (F4249)
- O2a2b1a2a1a3b2b1a (FGC23868)
- O2a2b1a2a1a3b2b2 (CTS335)
- O2a2b1a2a1a3b2b1 (F4249)
- O2a2b1a2a1a3b2a (L1360)
- O2a2b1a2a1a3a (F3607)
- O2a2b1a2a1a1 (F48)
- O2a2b1a2a1b (CTS53)
- O2a2b1a2a1b1 (CTS6373)
- O2a2b1a2a1b1a (A9473)
- O2a2b1a2a1b1 (CTS6373)
- O2a2b1a2a1c (F3386)
- O2a2b1a2a1d (Y29828)
- O2a2b1a2a1d1 (F735)
- O2a2b1a2a1d1a (FGC34973)
- O2a2b1a2a1d1b (F1739)
- O2a2b1a2a1d1 (F735)
- O2a2b1a2a1a (FGC16847/Z26091)
- O2a2b1a2a1 (F46/Y15)
- O2a2b1a2b (F743)
- O2a2b1a2b1 (CTS8481)
- O2a2b1a2b1a (CTS4325)
- O2a2b1a2b1a1 (A16629)
- O2a2b1a2b1a2 (CTS682)
- O2a2b1a2b1a (CTS4325)
- O2a2b1a2b2 (F748)
- O2a2b1a2b2a (F728)
- O2a2b1a2b1 (CTS8481)
- O2a2b1a2c (Page101)
- O2a2b1a2a (F79)
- O2a2b1a1 (M117/Page23)
- O2a2b1a (F450/M1667)
- O2a2b2 (AM01822/F3223)
- O2a2b2a (AM01856/F871)
- O2a2b2a1 (N7)
- O2a2b2a1a (F4110)
- O2a2b2a1a1 (F4068)
- O2a2b2a1a2 (SK1780)
- O2a2b2a1b (F4124)
- O2a2b2a1b1 (IMS-JST008425p6)
- O2a2b2a1b2 (BY15188)
- O2a2b2a1b2a (F16411)
- O2a2b2a1a (F4110)
- O2a2b2a2 (AM01845/F706)
- O2a2b2a2a (F717)
- O2a2b2a2a1 (F3612)
- O2a2b2a2a2 (SK1783)
- O2a2b2a2b (AM01847/B451)
- O2a2b2a2b1 (A17418)
- O2a2b2a2b2 (AM01756)
- O2a2b2a2b2a (B450)
- O2a2b2a2b2b (AM00472/B452)
- O2a2b2a2b2b1 (F18942)
- O2a2b2a2b2c (A16427)
- O2a2b2a2a (F717)
- O2a2b2a1 (N7)
- O2a2b2b (A16433)
- O2a2b2b1 (A16438)
- O2a2b2b1a (SK1775)
- O2a2b2b1a1 (SK1774)
- O2a2b2b1b (A16440)
- O2a2b2b1a (SK1775)
- O2a2b2b1 (A16438)
- O2a2b2a (AM01856/F871)
- O2a2b1 (M134)
- O2a2a (M188)
- O2a3 (M300)
- O2a4 (M333)
- O2a1 (L127.1)
- O2b (F742)
- O2b1 (F1150)
- O2b1a (F837)
- O2b1a1 (F1025)
- O2b1a (F837)
- O2b2 (F1055)
- O2b2a (F3021)
- O2b1 (F1150)
- O2a (M324)
- O1 (F265/M1354, CTS2866, F75/M1297, F429/M1415, F465/M1422)
See also
Genetics
Y-DNA O subclades
Y-DNA backbone tree
Notes
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Monika Karmin, Rodrigo Flores, Lauri Saag, Georgi Hudjashov, Nicolas Brucato, Chelzie Crenna-Darusallam, Maximilian Larena, Phillip L Endicott, Mattias Jakobsson, J Stephen Lansing, Herawati Sudoyo, Matthew Leavesley, Mait Metspalu, François-Xavier Ricaut, and Murray P Cox, "Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages," Molecular Biology and Evolution, Volume 39, Issue 3, March 2022, https://doi.org/10.1093/molbev/msac045
- ↑ 2.0 2.1 Poznik, G David; Xue, Yali; Mendez, Fernando L; Willems, Thomas F; Massaia, Andrea; Wilson Sayres, Melissa A; Ayub, Qasim; McCarthy, Shane A et al. (June 2016). "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences". Nature Genetics 48 (6): 593–599. doi:10.1038/ng.3559. PMID 27111036.
- ↑ ISOGG 2017
- ↑ 4.0 4.1 4.2 Ashirbekov, E. E.; Botbaev, D. M.; Belkozhaev, A. M.; Abayldaev, A. O.; Neupokoeva, A. S.; Mukhataev, J. E.; Alzhanuly, B.; Sharafutdinova, D. A. et al. (2017). "Distribution of Y-Chromosome Haplogroups of the Kazakh from the South Kazakhstan, Zhambyl, and Almaty Regions". Reports of the National Academy of Sciences of the Republic of Kazakhstan 6 (316): 85–95. http://rmebrk.kz/journals/3428/5747.pdf.
- ↑ "Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions". PLOS ONE 6 (3): e17548. March 2011. doi:10.1371/journal.pone.0017548. PMID 21412412. Bibcode: 2011PLoSO...617548D.
- ↑ 陆艳 [Lu Yan] (2011). 中国西部人群的遗传混合 [Genetic Mixture of Populations in Western China] (Thesis).
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 7.9 Karmin, Monika et al. (April 2015). "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research 25 (4): 459–466. doi:10.1101/gr.186684.114. PMID 25770088.
- ↑ 8.0 8.1 Yan, Shi; Wang, Chuan-Chao; Zheng, Hong-Xiang; Wang, Wei; Qin, Zhen-Dong; Wei, Lan-Hai; Wang, Yi; Pan, Xue-Dong et al. (29 August 2014). "Y Chromosomes of 40% Chinese Descend from Three Neolithic Super-Grandfathers". PLOS ONE 9 (8): e105691. doi:10.1371/journal.pone.0105691. PMID 25170956. Bibcode: 2014PLoSO...9j5691Y.
- ↑ Maxat Zhabagin, Zhaxylyk Sabitov, Pavel Tarlykov, Inkar Tazhigulova, Zukhra Junissova, Dauren Yerezhepov, Rakhmetolla Akilzhanov, Elena Zholdybayeva, Lan-Hai Wei, Ainur Akilzhanova, Oleg Balanovsky, and Elena Balanovska, "The medieval Mongolian roots of Y-chromosomal lineages from South Kazakhstan." BMC Genetics 2020, 21(Suppl 1):87. https://doi.org/10.1186/s12863-020-00897-5
- ↑ Shou, Wei-Hua; Qiao, En-Fa; Wei, Chuan-Yu; Dong, Yong-Li; Tan, Si-Jie; Shi, Hong; Tang, Wen-Ru; Xiao, Chun-Jie (May 2010). "Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians". Journal of Human Genetics 55 (5): 314–322. doi:10.1038/jhg.2010.30. PMID 20414255.
- ↑ Karafet, Tatiana M.; Mendez, Fernando L.; Sudoyo, Herawati; Lansing, J. Stephen; Hammer, Michael F. (March 2015). "Improved phylogenetic resolution and rapid diversification of Y-chromosome haplogroup K-M526 in Southeast Asia". European Journal of Human Genetics 23 (3): 369–373. doi:10.1038/ejhg.2014.106. PMID 24896152.
- ↑ "Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean". European Journal of Human Genetics 19 (1): 89–94. January 2011. doi:10.1038/ejhg.2010.128. PMID 20700146.
- ↑ 13.0 13.1 13.2 13.3 "The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages". American Journal of Human Genetics 76 (5): 894–901. May 2005. doi:10.1086/430051. PMID 15793703.
- ↑ "On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages". Molecular Biology and Evolution 26 (9): 2109–24. September 2009. doi:10.1093/molbev/msp120. PMID 19535740.
- ↑ "Y-chromosome diversity is inversely associated with language affiliation in paired Austronesian- and Papuan-speaking communities from Solomon Islands". American Journal of Human Biology 18 (1): 35–50. January 2006. doi:10.1002/ajhb.20459. PMID 16378340.
- ↑ "Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels". Journal of Human Genetics 58 (12): 804–11. December 2013. doi:10.1038/jhg.2013.108. PMID 24132124.
- ↑ "[Gene pool of Buryats: clinal variability and territorial subdivision based on data of Y-chromosome markers]". Genetika 50 (2): 203–13. February 2014. doi:10.1134/s1022795413110082. PMID 25711029.
- ↑ "Culture creates genetic structure in the Caucasus: autosomal, mitochondrial, and Y-chromosomal variation in Daghestan". BMC Genetics 9: 47. July 2008. doi:10.1186/1471-2156-9-47. PMID 18637195.
- ↑ Balanovsky, Oleg; Dibirova, Khadizhat; Dybo, Anna; Mudrak, Oleg; Frolova, Svetlana; Pocheshkhova, Elvira; Haber, Marc; Platt, Daniel et al. (1 October 2011). "Parallel Evolution of Genes and Languages in the Caucasus Region". Molecular Biology and Evolution 28 (10): 2905–2920. doi:10.1093/molbev/msr126. PMID 21571925.
- ↑ Haber, Marc; Platt, Daniel E.; Ashrafian Bonab, Maziar; Youhanna, Sonia C.; Soria-Hernanz, David F.; Martínez-Cruz, Begoña; Douaihy, Bouchra; Ghassibe-Sabbagh, Michella et al. (28 March 2012). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ 21.0 21.1 "Strong maternal Khoisan contribution to the South African coloured population: a case of gender-biased admixture". American Journal of Human Genetics 86 (4): 611–20. April 2010. doi:10.1016/j.ajhg.2010.02.014. PMID 20346436.
- ↑ Berniell-Lee, Gemma; Plaza, Stéphanie; Bosch, Elena; Calafell, Francesc; Jourdan, Eric; Césari, Maya; Lefranc, Gérard; Comas, David (May 2008). "Admixture and sexual bias in the population settlement of La Réunion Island (Indian Ocean)". American Journal of Physical Anthropology 136 (1): 100–107. doi:10.1002/ajpa.20783. PMID 18186507.
- ↑ "Y-chromosomal diversity in Haiti and Jamaica: contrasting levels of sex-biased gene flow". American Journal of Physical Anthropology 148 (4): 618–31. August 2012. doi:10.1002/ajpa.22090. PMID 22576450.
- ↑ "Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba". BMC Evolutionary Biology 8: 213. July 2008. doi:10.1186/1471-2148-8-213. PMID 18644108.
- ↑ Hammer, Michael F.; Chamberlain, Veronica F.; Kearney, Veronica F.; Stover, Daryn; Zhang, Gina; Karafet, Tatiana; Walsh, Bruce; Redd, Alan J. (December 2006). "Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases". Forensic Science International 164 (1): 45–55. doi:10.1016/j.forsciint.2005.11.013. PMID 16337103.
- ↑ Stefflova, Klara; Dulik, Matthew C.; Pai, Athma A.; Walker, Amy H.; Zeigler-Johnson, Charnita M.; Gueye, Serigne M.; Schurr, Theodore G.; Rebbeck, Timothy R. (25 November 2009). "Evaluation of Group Genetic Ancestry of Populations from Philadelphia and Dakar in the Context of Sex-Biased Admixture in the Americas". PLOS ONE 4 (11): e7842. doi:10.1371/journal.pone.0007842. PMID 19946364. Bibcode: 2009PLoSO...4.7842S.
- ↑ "Afghan Hindu Kush: where Eurasian sub-continent gene flows converge". PLOS ONE 8 (10): e76748. 2013. doi:10.1371/journal.pone.0076748. PMID 24204668. Bibcode: 2013PLoSO...876748D.
- ↑ Magoon, Gregory R.; Banks, Raymond H.; Rottensteiner, Christian; Schrack, Bonnie E.; Tilroe, Vincent O.; Robb, Terry; Grierson, Andrew J. (13 December 2013). "Generation of high-resolution a priori Y-chromosome phylogenies using 'next-generation' sequencing data". bioRxiv 10.1101/000802.Template:S2cid
- ↑ 29.000 29.001 29.002 29.003 29.004 29.005 29.006 29.007 29.008 29.009 29.010 29.011 29.012 29.013 29.014 29.015 29.016 29.017 29.018 29.019 29.020 29.021 29.022 29.023 29.024 29.025 29.026 29.027 29.028 29.029 29.030 29.031 29.032 29.033 29.034 29.035 29.036 29.037 29.038 29.039 29.040 29.041 29.042 29.043 29.044 29.045 29.046 29.047 29.048 29.049 29.050 29.051 29.052 29.053 29.054 29.055 29.056 29.057 29.058 29.059 29.060 29.061 29.062 29.063 29.064 29.065 29.066 29.067 29.068 29.069 29.070 29.071 29.072 29.073 29.074 29.075 29.076 29.077 29.078 29.079 29.080 29.081 29.082 29.083 29.084 29.085 29.086 29.087 29.088 29.089 29.090 29.091 29.092 29.093 29.094 29.095 29.096 29.097 29.098 29.099 29.100 29.101 29.102 29.103 29.104 29.105 29.106 29.107 29.108 29.109 29.110 29.111 29.112 29.113 29.114 29.115 29.116 29.117 29.118 29.119 29.120 29.121 29.122 29.123 29.124 29.125 29.126 29.127 29.128 29.129 29.130 29.131 29.132 29.133 29.134 29.135 29.136 29.137 29.138 29.139 29.140 29.141 29.142 29.143 29.144 29.145 29.146 29.147 29.148 29.149 29.150 29.151 29.152 29.153 29.154 29.155 29.156 29.157 29.158 29.159 29.160 29.161 29.162 29.163 29.164 29.165 29.166 29.167 29.168 29.169 29.170 29.171 29.172 29.173 29.174 29.175 29.176 29.177 29.178 29.179 29.180 29.181 29.182 29.183 29.184 29.185 29.186 29.187 29.188 29.189 29.190 29.191 29.192 29.193 29.194 29.195 29.196 29.197 29.198 29.199 29.200 29.201 29.202 29.203 29.204 29.205 29.206 29.207 29.208 29.209 29.210 29.211 29.212 29.213 29.214 29.215 29.216 29.217 29.218 29.219 29.220 29.221 29.222 29.223 29.224 29.225 29.226 29.227 29.228 29.229 29.230 29.231 29.232 29.233 29.234 29.235 29.236 29.237 29.238 29.239 29.240 29.241 29.242 29.243 29.244 29.245 29.246 29.247 29.248 29.249 29.250 29.251 29.252 29.253 29.254 29.255 29.256 29.257 29.258 29.259 29.260 29.261 29.262 29.263 29.264 29.265 29.266 29.267 29.268 29.269 29.270 29.271 29.272 29.273 29.274 29.275 29.276 29.277 29.278 29.279 29.280 29.281 29.282 29.283 29.284 29.285 29.286 29.287 29.288 29.289 29.290 29.291 29.292 29.293 29.294 29.295 29.296 29.297 29.298 29.299 29.300 29.301 29.302 29.303 29.304 29.305 29.306 29.307 29.308 29.309 29.310 29.311 29.312 29.313 29.314 29.315 29.316 29.317 29.318 29.319 29.320 29.321 29.322 29.323 29.324 29.325 29.326 29.327 29.328 29.329 29.330 29.331 29.332 29.333 29.334 29.335 29.336 29.337 29.338 29.339 29.340 29.341 29.342 29.343 29.344 29.345 29.346 29.347 29.348 29.349 29.350 29.351 29.352 29.353 29.354 29.355 29.356 29.357 29.358 29.359 29.360 29.361 29.362 29.363 29.364 29.365 29.366 29.367 29.368 29.369 29.370 29.371 29.372 29.373 29.374 29.375 29.376 29.377 29.378 29.379 29.380 29.381 29.382 29.383 29.384 29.385 29.386 29.387 29.388 29.389 29.390 29.391 29.392 29.393 29.394 29.395 29.396 29.397 29.398 29.399 29.400 29.401 29.402 29.403 29.404 29.405 29.406 29.407 29.408 29.409 29.410 29.411 29.412 29.413 29.414 29.415 29.416 29.417 29.418 29.419 29.420 29.421 29.422 29.423 29.424 29.425 29.426 29.427 29.428 29.429 29.430 29.431 29.432 29.433 29.434 29.435 29.436 29.437 29.438 29.439 29.440 29.441 29.442 29.443 29.444 29.445 29.446 29.447 29.448 29.449 29.450 29.451 29.452 YFull Haplogroup YTree v5.04 at 16 May 2017
- ↑ 30.000 30.001 30.002 30.003 30.004 30.005 30.006 30.007 30.008 30.009 30.010 30.011 30.012 30.013 30.014 30.015 30.016 30.017 30.018 30.019 30.020 30.021 30.022 30.023 30.024 30.025 30.026 30.027 30.028 30.029 30.030 30.031 30.032 30.033 30.034 30.035 30.036 30.037 30.038 30.039 30.040 30.041 30.042 30.043 30.044 30.045 30.046 30.047 30.048 30.049 30.050 30.051 30.052 30.053 30.054 30.055 30.056 30.057 30.058 30.059 30.060 30.061 30.062 30.063 30.064 30.065 30.066 30.067 30.068 30.069 30.070 30.071 30.072 30.073 30.074 30.075 30.076 30.077 30.078 30.079 30.080 30.081 30.082 30.083 30.084 30.085 30.086 30.087 30.088 30.089 30.090 30.091 30.092 30.093 30.094 30.095 30.096 30.097 30.098 30.099 30.100 30.101 30.102 30.103 30.104 30.105 30.106 30.107 30.108 30.109 30.110 30.111 30.112 30.113 30.114 30.115 30.116 30.117 30.118 30.119 30.120 30.121 30.122 30.123 30.124 30.125 30.126 30.127 30.128 30.129 30.130 30.131 30.132 30.133 30.134 30.135 30.136 30.137 30.138 30.139 30.140 30.141 30.142 30.143 30.144 30.145 30.146 Phylogenetic tree of Haplogroup O at 23mofang
- ↑ 31.00 31.01 31.02 31.03 31.04 31.05 31.06 31.07 31.08 31.09 31.10 31.11 31.12 31.13 31.14 31.15 31.16 31.17 31.18 31.19 31.20 31.21 31.22 31.23 31.24 31.25 31.26 31.27 31.28 31.29 31.30 31.31 31.32 31.33 31.34 31.35 31.36 31.37 31.38 31.39 31.40 31.41 Phylogenetic tree of Haplogroup O-F175 at TheYtree
- ↑ 32.0 32.1 "An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4". European Journal of Human Genetics 19 (9): 1013–5. September 2011. doi:10.1038/ejhg.2011.64. PMID 21505448.
- ↑ 33.00 33.01 33.02 33.03 33.04 33.05 33.06 33.07 33.08 33.09 33.10 33.11 33.12 33.13 33.14 33.15 33.16 33.17 33.18 33.19 33.20 33.21 33.22 33.23 33.24 33.25 33.26 33.27 33.28 33.29 33.30 33.31 33.32 33.33 33.34 33.35 33.36 33.37 33.38 33.39 33.40 33.41 Macholdt, Enrico; Arias, Leonardo; Duong, Nguyen Thuy; Ton, Nguyen Dang; Van Phong, Nguyen; Schröder, Roland; Pakendorf, Brigitte; Van Hai, Nong et al. (May 2020). "The paternal and maternal genetic history of Vietnamese populations". European Journal of Human Genetics 28 (5): 636–645. doi:10.1038/s41431-019-0557-4. PMID 31827276.
- ↑ 34.0 34.1 O Y-Haplogroup Project at Family Tree DNA
- ↑ 35.00 35.01 35.02 35.03 35.04 35.05 35.06 35.07 35.08 35.09 35.10 35.11 35.12 35.13 35.14 35.15 35.16 35.17 35.18 35.19 35.20 35.21 35.22 35.23 35.24 35.25 35.26 35.27 35.28 35.29 35.30 35.31 Y-DNA Haplotree at Family Tree DNA
- ↑ 36.0 36.1 36.2 Damgaard, Peter de Barros; Marchi, Nina; Rasmussen, Simon; Peyrot, Michaël; Renaud, Gabriel; Korneliussen, Thorfinn; Moreno-Mayar, J. Víctor; Pedersen, Mikkel Winther et al. (May 2018). "137 ancient human genomes from across the Eurasian steppes". Nature 557 (7705): 369–374. doi:10.1038/s41586-018-0094-2. PMID 29743675. Bibcode: 2018Natur.557..369D.
- ↑ 37.0 37.1 37.2 37.3 37.4 37.5 37.6 37.7 37.8 Zhang, Xiaoming; Kampuansai, Jatupol; Qi, Xuebin; Yan, Shi; Yang, Zhaohui; Serey, Bun; Sovannary, Tuot; Bunnath, Long et al. (27 June 2014). "An Updated Phylogeny of the Human Y-Chromosome Lineage O2a-M95 with Novel SNPs". PLOS ONE 9 (6): e101020. doi:10.1371/journal.pone.0101020. PMID 24972021. Bibcode: 2014PLoSO...9j1020Z.
- ↑ 38.0 38.1 Lippold, Sebastian; Xu, Hongyang; Ko, Albert; Li, Mingkun; Renaud, Gabriel; Butthof, Anne; Schröder, Roland; Stoneking, Mark (December 2014). "Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences". Investigative Genetics 5 (1): 13. doi:10.1186/2041-2223-5-13. PMID 25254093.
- ↑ 39.000 39.001 39.002 39.003 39.004 39.005 39.006 39.007 39.008 39.009 39.010 39.011 39.012 39.013 39.014 39.015 39.016 39.017 39.018 39.019 39.020 39.021 39.022 39.023 39.024 39.025 39.026 39.027 39.028 39.029 39.030 39.031 39.032 39.033 39.034 39.035 39.036 39.037 39.038 39.039 39.040 39.041 39.042 39.043 39.044 39.045 39.046 39.047 39.048 39.049 39.050 39.051 39.052 39.053 39.054 39.055 39.056 39.057 39.058 39.059 39.060 39.061 39.062 39.063 39.064 39.065 39.066 39.067 39.068 39.069 39.070 39.071 39.072 39.073 39.074 39.075 39.076 39.077 39.078 39.079 39.080 39.081 39.082 39.083 39.084 39.085 39.086 39.087 39.088 39.089 39.090 39.091 39.092 39.093 39.094 39.095 39.096 39.097 39.098 39.099 39.100 39.101 Kutanan, Wibhu; Kampuansai, Jatupol; Srikummool, Metawee; Brunelli, Andrea; Ghirotto, Silvia; Arias, Leonardo; Macholdt, Enrico; Hübner, Alexander et al. (1 July 2019). "Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations". Molecular Biology and Evolution 36 (7): 1490–1506. doi:10.1093/molbev/msz083. PMID 30980085.
- ↑ Xia, Zi-Yang; Yan, Shi; Wang, Chuan-Chao; Zheng, Hong-Xiang; Zhang, Fan; Liu, Yu-Chi; Yu, Ge; Yu, Bin-Xia; Shu, Li-Li; Jin, Li (9 August 2019). "Inland-coastal bifurcation of southern East Asians revealed by Hmong-Mien genomic history". bioRxiv 10.1101/730903.Template:S2cid
- ↑ 41.0 41.1 41.2 41.3 41.4 41.5 41.6 41.7 Karafet, T. M.; Hallmark, B.; Cox, M. P.; Sudoyo, H.; Downey, S.; Lansing, J. S.; Hammer, M. F. (1 August 2010). "Major East-West Division Underlies Y Chromosome Stratification across Indonesia". Molecular Biology and Evolution 27 (8): 1833–1844. doi:10.1093/molbev/msq063. PMID 20207712.
- ↑ 42.0 42.1 42.2 42.3 42.4 42.5 42.6 42.7 42.8 Trejaut, Jean A; Poloni, Estella S; Yen, Ju-Chen; Lai, Ying-Hui; Loo, Jun-Hun; Lee, Chien-Liang; He, Chun-Lin; Lin, Marie (2014). "Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia". BMC Genetics 15 (1): 77. doi:10.1186/1471-2156-15-77. PMID 24965575.
- ↑ 43.0 43.1 43.2 43.3 43.4 43.5 43.6 Xue, Yali; Zerjal, Tatiana; Bao, Weidong; Zhu, Suling; Shu, Qunfang; Xu, Jiujin; Du, Ruofu; Fu, Songbin et al. (1 April 2006). "Male Demography in East Asia: A North–South Contrast in Human Population Expansion Times". Genetics 172 (4): 2431–2439. doi:10.1534/genetics.105.054270. PMID 16489223.
- ↑ 44.0 44.1 44.2 Hammer, Michael F.; Karafet, Tatiana M.; Park, Hwayong; Omoto, Keiichi; Harihara, Shinji; Stoneking, Mark; Horai, Satoshi (January 2006). "Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes". Journal of Human Genetics 51 (1): 47–58. doi:10.1007/s10038-005-0322-0. PMID 16328082.
- ↑ 45.0 45.1 45.2 45.3 45.4 45.5 Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan; Zhang, Weiling; Akey, Joshua; Huang, Wei; Shen, Di et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- ↑ Jing, Chen; Hui, Li; Zhen-Dong, Qin; Wen-Hong, Liu; Wei-Xiong, Lin; Rui-Xing, Yin; Li, Jin; Shang-Ling, Pan (December 2006). "Y-chromosome Genotyping and Genetic Structure of Zhuang Populations". Acta Genetica Sinica 33 (12): 1060–1072. doi:10.1016/S0379-4172(06)60143-1. PMID 17185165.
- ↑ 47.0 47.1 47.2 He, Jun-Dong; Peng, Min-Sheng; Quang, Huy Ho; Dang, Khoa Pham; Trieu, An Vu; Wu, Shi-Fang; Jin, Jie-Qiong; Murphy, Robert W. et al. (7 May 2012). "Patrilineal Perspective on the Austronesian Diffusion in Mainland Southeast Asia". PLOS ONE 7 (5): e36437. doi:10.1371/journal.pone.0036437. PMID 22586471. Bibcode: 2012PLoSO...736437H.
- ↑ 48.0 48.1 48.2 Bing Su, Li Jin, Peter Underhill, et al. (2000), "Polynesian origins: Insights from the Y chromosome." PNAS, vol. 97, no. 15, 8225–8228.
- ↑ 49.0 49.1 49.2 49.3 49.4 49.5 Cai, Xiaoyun; Qin, Zhendong; Wen, Bo; Xu, Shuhua; Wang, Yi; Lu, Yan; Wei, Lanhai; Wang, Chuanchao et al. (31 August 2011). "Human Migration through Bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum Revealed by Y Chromosomes". PLOS ONE 6 (8): e24282. doi:10.1371/journal.pone.0024282. PMID 21904623. Bibcode: 2011PLoSO...624282C.
- ↑ Wen, Bo (2004). "The origin of Mosuo people as revealed by mtDNA and Y chromosome variation". Science in China Series C 47 (1): 1–10. doi:10.1360/02yc0207. PMID 15382670.
- ↑ 51.0 51.1 Peng, Min-Sheng; He, Jun-Dong; Fan, Long; Liu, Jie; Adeola, Adeniyi C; Wu, Shi-Fang; Murphy, Robert W; Yao, Yong-Gang et al. (August 2014). "Retrieving Y chromosomal haplogroup trees using GWAS data". European Journal of Human Genetics 22 (8): 1046–1050. doi:10.1038/ejhg.2013.272. PMID 24281365.
- ↑ 52.00 52.01 52.02 52.03 52.04 52.05 52.06 52.07 52.08 52.09 52.10 52.11 52.12 52.13 52.14 52.15 52.16 52.17 52.18 52.19 52.20 52.21 52.22 52.23 52.24 52.25 52.26 52.27 "Improved phylogenetic resolution for Y-chromosome Haplogroup O2a1c-002611". Scientific Reports 7 (1): 1146. April 2017. doi:10.1038/s41598-017-01340-z. PMID 28442769. Bibcode: 2017NatSR...7.1146Y.
- ↑ "O-M159单倍群详情". https://www.23mofang.com/ancestry/ytree/O-M159/detail.
- ↑ "O-Mf22947单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF22947/detail.
- ↑ "O-Z25518单倍群详情". https://www.23mofang.com/ancestry/ytree/O-Z25518/detail.
- ↑ "O-Mf14135单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF14135/detail.
- ↑ "夏代东部大族祖源分析-23魔方祖源基因检测". https://www.23mofang.com/ancestry/family/626508f6d6cf927a104d0a0e.
- ↑ 58.00 58.01 58.02 58.03 58.04 58.05 58.06 58.07 58.08 58.09 58.10 58.11 58.12 58.13 58.14 58.15 58.16 58.17 58.18 58.19 58.20 58.21 58.22 58.23 58.24 58.25 58.26 58.27 58.28 58.29 58.30 58.31 58.32 Time Tree of Y-DNA haplogroup O-M175 at FamilyTreeDNA Discover
- ↑ Li, Hui; Huang, Ying; Mustavich, Laura F.; Zhang, Fan; Tan, Jing-Ze; Wang, Ling-E; Qian, Ji; Gao, Meng-He et al. (November 2007). "Y chromosomes of prehistoric people along the Yangtze River". Human Genetics 122 (3–4): 383–388. doi:10.1007/s00439-007-0407-2. PMID 17657509.
- ↑ "O-Mf9858单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF9858/detail.
- ↑ "O-F1262单倍群详情". https://www.23mofang.com/ancestry/ytree/O-F1262/detail.
- ↑ "O-Mf106428单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF106428/detail.
- ↑ 63.0 63.1 63.2 63.3 63.4 63.5 Kutanan, Wibhu; Shoocongdej, Rasmi; Srikummool, Metawee; Hübner, Alexander; Suttipai, Thanatip; Srithawong, Suparat; Kampuansai, Jatupol; Stoneking, Mark (November 2020). "Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand". European Journal of Human Genetics 28 (11): 1563–1579. doi:10.1038/s41431-020-0693-x. PMID 32690935.
- ↑ "O-Mf48275单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF48275/detail.
- ↑ "O-F14832单倍群详情". https://www.23mofang.com/ancestry/ytree/O-F14832/detail.
- ↑ "O-Y140772单倍群详情". https://www.23mofang.com/ancestry/ytree/O-Y140772/detail.
- ↑ "O-Z25400单倍群详情". https://www.23mofang.com/ancestry/ytree/O-Z25400/detail.
- ↑ "O-Mf36985单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF36985/detail.
- ↑ "O-Mf193618单倍群详情". https://www.23mofang.com/ancestry/ytree/O-MF193618/detail.
- ↑ Huang, Xiufeng; Xia, Zi-Yang; Bin, Xiaoyun; He, Guanglin; Guo, Jianxin (30 June 2022). "Genomic Insights Into the Demographic History of the Southern Chinese". Frontiers in Ecology and Evolution (Frontiers Media SA) 10. doi:10.3389/fevo.2022.853391. ISSN 2296-701X.
- ↑ "Assignment of Y-chromosomal SNPs found in Japanese population to Y-chromosomal haplogroup tree". Journal of Human Genetics 58 (4): 195–201. April 2013. doi:10.1038/jhg.2012.159. PMID 23389242.
- ↑ Wei, Lan-Hai; Yan, Shi; Teo, Yik-Ying; Huang, Yun-Zhi; Wang, Ling-Xiang; Yu, Ge; Saw, Woei-Yuh; Ong, Rick Twee-Hee et al. (5 April 2017). "Phylogeography of Y-chromosome haplogroup O3a2b2-N6 reveals patrilineal traces of Austronesian populations on the eastern coastal regions of Asia". PLOS ONE 12 (4): e0175080. doi:10.1371/journal.pone.0175080. PMID 28380021. Bibcode: 2017PLoSO..1275080W.
- ↑ 73.0 73.1 73.2 73.3 Mondal, Mayukh; Casals, Ferran; Xu, Tina; Dall'Olio, Giovanni M; Pybus, Marc; Netea, Mihai G; Comas, David; Laayouni, Hafid et al. (September 2016). "Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation". Nature Genetics 48 (9): 1066–1070. doi:10.1038/ng.3621. PMID 27455350.
- ↑ "Y-DNA Haplogroup D-M174 and its Subclades - 2017". http://www.isogg.org/tree/ISOGG_HapgrpD.html.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
Further reading
- Jimmy G. Harris, Somsonge Burusphat and James E. Harris, ed (2007). "The power of language over the past: Tai settlement and Tai linguistics in southern China and northern Vietnam". Studies in Southeast Asian Languages and Linguistics (Bangkok, Thailand: Ek Phim Thai Co. Ltd.). http://ling.uta.edu/~jerry/pol.pdf.
- van Driem, George (2011). "Rice and the Austroasiatic and Hmong-Mien Homelands". in Enfield, N. J. Dynamics of Human Diversity. Pacific Linguistics. pp. 361–390. ISBN 978-0-85883-638-9. https://www.himalayanlanguages.org/files/driem/pdfs/2011Rice%20and%20the%20Austroasiatic%20and%20Hmong-Mien%20homelands.pdf.
- "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics 15 (1): 121–6. January 2007. doi:10.1038/sj.ejhg.5201726. PMID 17047675.
- Fornarino, Simona; Pala, Maria; Battaglia, Vincenza; Maranta, Ramona; Achilli, Alessandro; Modiano, Guido; Torroni, Antonio; Semino, Ornella et al. (December 2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation". BMC Evolutionary Biology 9 (1): 154. doi:10.1186/1471-2148-9-154. PMID 19573232.
- "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. May 2008. doi:10.1101/gr.7172008. PMID 18385274.
- "The impact of the Austronesian expansion: evidence from mtDNA and Y chromosome diversity in the Admiralty Islands of Melanesia". Molecular Biology and Evolution 25 (7): 1362–74. July 2008. doi:10.1093/molbev/msn078. PMID 18390477.
- Kharkov, V. N.; Stepanov, V. A.; Medvedeva, O. F.; Spiridonova, M. G.; Voevoda, M. I.; Tadinova, V. N.; Puzyrev, V. P. (2007). "Gene pool differences between Northern and Southern Altaians inferred from the data on Y-chromosomal haplogroups". Russian Journal of Genetics 43 (5): 551–562. doi:10.1134/S1022795407050110. PMID 17633562.
- "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea". Investigative Genetics 2 (1): 10. April 2011. doi:10.1186/2041-2223-2-10. PMID 21463511.
- "Ho Ne (She) is Hmongic: One final argument". Linguistics of the Tibeto-Burman Area 21 (2): 97–109. 1998. http://sealang.net/sala/archives/pdf8/ratliff1998ho.pdf.
- "A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe". European Journal of Human Genetics 15 (2): 204–11. February 2007. doi:10.1038/sj.ejhg.5201748. PMID 17149388.
- "Unexpected NRY chromosome variation in Northern Island Melanesia". Molecular Biology and Evolution 23 (8): 1628–41. August 2006. doi:10.1093/molbev/msl028. PMID 16754639.
- "Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122". American Journal of Human Genetics 77 (3): 408–19. September 2005. doi:10.1086/444436. PMID 16080116.
- "Polynesian origins: insights from the Y chromosome". Proceedings of the National Academy of Sciences of the United States of America 97 (15): 8225–8. July 2000. doi:10.1073/pnas.97.15.8225. PMID 10899994. Bibcode: 2000PNAS...97.8225S.
- "The Eurasian heartland: a continental perspective on Y-chromosome diversity". Proceedings of the National Academy of Sciences of the United States of America 98 (18): 10244–9. August 2001. doi:10.1073/pnas.171305098. PMID 11526236. Bibcode: 2001PNAS...9810244W.
- "The origin of Mosuo people as revealed by mtDNA and Y chromosome variation". Science in China Series C: Life Sciences 47 (1): 1–10. February 2004. doi:10.1360/02yc0207. PMID 15382670.
- "Male demography in East Asia: a north-south contrast in human population expansion times". Genetics 172 (4): 2431–9. April 2006. doi:10.1534/genetics.105.054270. PMID 16489223.
External links
- Bradshaw Foundation. "Journey of Man - The Peopling of the World". http://www.bradshawfoundation.com/journey/.
- ISOGG (2012). "Y-DNA Haplogroup O and its Subclades - 2012". http://www.isogg.org/tree/ISOGG_HapgrpO.html.
- TMC (1998). "Genetic Findings Support 'Out of Africa' Theory". http://www.tmc.edu/tmcnews/10_15_98/page_08.html.
- Spread of Haplogroup O, from The Genographic Project, National Geographic
- Y-DNA Phylogenetic Tree of Haplogroup O (DNAHaplogroups.org)
- Migration patterns of early Humans and the full size map
- China DNA at Family Tree DNA
 |

