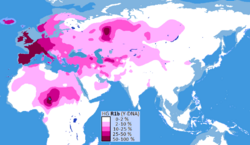
Biology:Haplogroup R1b
| Haplogroup R1b | |
|---|---|
 | |
| Possible time of origin | Probably soon after R1, possibly between 18,000-14,000 BC[1] |
| Possible place of origin | Western Asia, North Eurasia or Eastern Europe[2] |
| Ancestor | R1 |
| Descendants |
|
| Defining mutations | M343 |
Haplogroup R1b (R-M343), previously known as Hg1 and Eu18, is a human Y-chromosome haplogroup.
It is the most frequently occurring paternal lineage in Western Europe, as well as some parts of Russia (e.g. the Bashkirs) and across the Sahel in Central Africa, namely: Cameroon, Chad, Guinea Mauritania, Mali, Niger, Nigeria and Senegal (concentrated in parts of Chad with concentration in the Hausa Tribe and among the Chadic-speaking ethnic groups of Cameroon).
The clade is also present at lower frequencies throughout Eastern Europe, Western Asia, Central Asia as well as parts of North Africa, South Asia and Central Asia.
R1b has two primary branches: R1b1-L754 and R1b2-PH155. R1b1-L754 has two major subclades: R1b1a1b-M269, which predominates in Western Europe, and R1b1b-V88, which is today common in parts of Central Africa. The other branch, R1b2-PH155, is so rare and widely dispersed that it is difficult to draw any conclusions about its origins. It has been found in Bahrain, India , Nepal, Bhutan, Ladakh, Tajikistan, Turkey, and Western China.
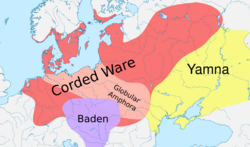
According to ancient DNA studies, most R1a and R1b lineages would have expanded from the Pontic Steppe along with the Indo-European languages.[2][3][4][5][6]
Origin and dispersal
The age of R1 was estimated by Tatiana Karafet et al. (2008) at between 12,500 and 25,700 BP, and most probably occurred about 18,500 years ago.[7] Since the earliest known example has been dated at circa 14,000 BP, and belongs to R1b1 (R-L754),[1] R1b must have arisen relatively soon after the emergence of R1.
Early human remains found to carry R1b include:
- Villabruna 1 (individual I9030), a Western Hunter-Gatherer (WHG), found in an Epigravettian culture setting in the Cismon valley (modern Veneto, Italy), who lived circa 14000 BP and belonged to R1b1a.[1][8]
- Several males of the Iron Gates Mesolithic in the Balkans buried between 11200 and 8200 BP carried R1b1a1a. These individuals were determined to be largely of WHG ancestry, with slight Eastern Hunter-Gatherer (EHG) admixture.[9]
- Several males of the Mesolithic Kunda culture and Neolithic Narva culture buried in the Zvejnieki burial ground in modern-day Latvia c. 9500–6000 BP carried R1b1b.[10][11] These individuals were determined to be largely of WHG ancestry, with slight EHG admixture.[10]
- Several Mesolithic and Neolithic males buried at Deriivka and Vasil'evka in modern-day Ukraine c. 9500-7000 BP carried R1b1a.[12] These individuals were largely of EHG ancestry, with significant WHG admixture.[13]
- A WHG male buried at Ostrovul Corbuli, Romania c. 8700 BP carried R1b1c.[14][15][16]
- A male buried at Lepenski Vir, Serbia c. 8200-7900 BP carried R1b1a.[17]
- An EHG buried near Samara, Russia 7500 BP carried R1b1a1a.[18]
- An Eneolithic male buried at Khvalynsk, Russia c. 7200-6000 BP carried R1b1a.[19]
- A Neolithic male buried at Els Trocs, Spain c. 7178-7066 BP, who may have belonged to the Epi-Cardial culture,[20] was found to be a carrier of R1b1.[21][22][23]
- A Late Chalcolithic male buried in Smyadovo, Bulgaria c. 6500 BP carried R1b1a.[24]
- An Early Copper Age male buried in Cannas di Sotto, Carbonia, Sardinia c. 6450 BP carried R1b1b2.[25]
- A male of the Baalberge group in Central Europe buried c. 5600 BP carried R1b1a.[26]
- A male of the Botai culture in Central Asia buried c. 5500 BP carried R1b1a1 (R1b-M478).[27]
- 7 males that were tested of the Yamnaya culture were all found to belong to the M269 subclade of haplogroup R1b.[28]
R1b is a subclade within the "macro-haplogroup" K (M9), the most common group of human male lines outside of Africa. K is believed to have originated in Asia (as is the case with an even earlier ancestral haplogroup, F (F-M89). Karafet T. et al. (2014) "rapid diversification process of K-M526 likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q".[29]
Three genetic studies in 2015 gave support to the Kurgan hypothesis of Marija Gimbutas regarding the Proto-Indo-European homeland. According to those studies, haplogroups R1b-M269 and R1a, now the most common in Europe (R1a is also common in South Asia) would have expanded from the West Eurasian Steppe, along with the Indo-European languages; they also detected an autosomal component present in modern Europeans which was not present in Neolithic Europeans, which would have been introduced with paternal lineages R1b and R1a, as well as Indo-European languages.[2][3][4]

Analysis of ancient Y-DNA from the remains from early Neolithic Central and North European Linear Pottery culture settlements have not yet found males belonging to haplogroup R1b-M269.[30][31] Olalde et al. (2017) trace the spread of haplogroup R1b-M269 in western Europe, particularly Britain, to the spread of the Beaker culture, with a sudden appearance of many R1b-M269 haplogroups in Western Europe ca. 5000–4500 years BP during the early Bronze Age.[32]
Structure
External phylogeny of R1b
The broader haplogroup R (M207) is a primary subclade of haplogroup P1 (M45) itself a primary branch of P (P295), which is also known as haplogroup K2b2. R-M207 is therefore a secondary branch of K2b (P331), and a direct descendant of K2 (M526).
There was "an initial rapid diversification" of K-M526, according to Karafet et al. (2014), which "likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q".
- Phylogeny within K2b
- P P295/PF5866/S8 (also known as K2b2).
Internal structure of R1b
Names such as R1b, R1b1 and so on are phylogenetic (i.e. "family tree") names which make clear their place within the branching of haplogroups, or the phylogenetic tree. An alternative way of naming the same haplogroups and subclades refers to their defining SNP mutations: for example, R-M343 is equivalent to R1b.[33] Phylogenetic names change with new discoveries and SNP-based names are consequently reclassified within the phylogenetic tree. In some cases, an SNP is found to be unreliable as a defining mutation and an SNP-based name is removed completely. For example, before 2005, R1b was synonymous with R-P25, which was later reclassified as R1b1; in 2016, R-P25 was removed completely as a defining SNP, due to a significant rate of back-mutation.[34] (Below is the basic outline of R1b according to the ISOGG Tree as it stood on January 30, 2017.[35])
|
Geographical distribution
R1b* (R-M343*)
No confirmed cases of R1b* (R-M343*) – that is R1b (xR1b1, R1b2), also known as R-M343 (xL754, PH155) – have been reported in peer-reviewed literature.
- R-M343 (xM73, M269, V88)
In early research, because R-M269, R-M73 and R-V88 are by far the most common forms of R1b, examples of R1b (xM73, xM269) were sometimes assumed to signify basal examples of "R1b*".[34] However, while the paragroup R-M343 (xM73, M269, V88) is rare, it does not preclude membership of rare and/or subsequently-discovered, relatively basal subclades of R1b, such as R-L278* (R1b*), R-L389* (R1b1a*), R-P297* (R1b1a1*), R-V1636 (R1b1a2) or R-PH155 (R1b2).
The population believed to have the highest proportion of R-M343 (xM73, M269, V88) are the Kurds of southeastern Kazakhstan with 13%.[40] However, more recently, a large study of Y-chromosome variation in Iran, revealed R-M343 (xV88, M73, M269) as high as 4.3% among Iranian sub-populations.[41]
It remains a possibility that some, or even most of these cases, may be R-L278* (R1b*), R-L389* (R1b1a*), R-P297* (R1b1a1*), R-V1636 (R1b1a2), R-PH155 (R1b2), R1b* (R-M343*), R1a* (R-M420*), an otherwise undocumented branch of R1 (R-M173), and/or back-mutations of a marker, from a positive to a negative ancestral state,[42] and hence constitute undocumented subclades of R1b.
A compilation of previous studies regarding the distribution of R1b can be found in Cruciani et al. (2010).[43] It is summarised in the table following. (Cruciani did not include some studies suggesting even higher frequencies of R1b1a1b [R-M269] in some parts of Western Europe.)
| Continent | Region | Sample size |
Total R1b | R-P25 (unreliable marker for R1b1*) |
R-V88 (R1b1b) |
R-M269 (R1b1a1a2) |
R-M73 (R1b1a1a1) |
|---|---|---|---|---|---|---|---|
| Africa | Northern Africa | 691 | 5.9% | 0.0% | 5.2% | 0.7% | 0.0% |
| Africa | Central Sahel Region | 461 | 23.0% | 0.0% | 23.0% | 0.0% | 0.0% |
| Africa | Western Africa | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Eastern Africa | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Southern Africa | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Europe | Western Europeans | 465 | 57.8% | 0.0% | 0.0% | 57.8% | 0.0% |
| Europe | North-west Europeans | 43 | 55.8% | 0.0% | 0.0% | 55.8% | 0.0% |
| Europe | Central Europeans | 77 | 42.9% | 0.0% | 0.0% | 42.9% | 0.0% |
| Europe | North Eastern Europeans | 74 | 1.4% | 0.0% | 0.0% | 1.4% | 0.0% |
| Europe | Russians | 60 | 6.7% | 0.0% | 0.0% | 6.7% | 0.0% |
| Europe | Eastern Europeans | 149 | 20.8% | 0.0% | 0.0% | 20.8% | 0.0% |
| Europe | South-east Europeans | 510 | 13.1% | 0.0% | 0.2% | 12.9% | 0.0% |
| Asia | West Asians | 328 | 5.8% | 0.0% | 0.3% | 5.5% | 0.0% |
| Asia | South Asians | 288 | 4.8% | 0.0% | 0.0% | 1.7% | 3.1% |
| Asia | South-east Asians | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | North-east Asians | 30 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | East Asians | 156 | 0.6% | 0.0% | 0.0% | 0.6% | 0.0% |
| Total | 5326 | ||||||
R1b (R-L278)
R-L278 among modern men falls into the R-L754 and R-PH155 subclades, though it is possible some very rare R-L278* may exist as not all examples have been tested for both branches. Examples may also exist in ancient DNA, though due to poor quality it is often impossible to tell whether or not the ancients carried the mutations that define subclades.
Some examples described in older articles, for example two found in Turkey,[33] are now thought to be mostly in the more recently discovered sub-clade R1b1b (R-V88). Most examples of R1b therefore fall into subclades R1b1b (R-V88) or R1b1a (R-P297). Cruciani et al. in the large 2010 study found 3 cases amongst 1173 Italians, 1 out of 328 West Asians and 1 out of 156 East Asians.[43] Varzari found 3 cases in Ukraine , in a study of 322 people from the Dniester–Carpathian Mountains region, who were P25 positive, but M269 negative.[44] Cases from older studies are mainly from Africa, the Middle East or Mediterranean, and are discussed below as probable cases of R1b1b (R-V88).
R1b1 (R-L754)
R-L754 contains the vast majority of R1b. The only known example of R-L754* (xL389, V88) is also the earliest known individual to carry R1b: "Villabruna 1", who lived circa 14,000 years BP (north east Italy). Villabruna 1 belonged to the Epigravettian culture.
R1b1a (R-L389)
R-L389, also known as R1b1a (L388/PF6468, L389/PF6531), contains the very common subclade R-P297 and the rare subclade R-V1636. It is unknown whether all previously reported R-L389* (xP297) belong to R-V1636 or not.
R1b1a1 (R-P297)
The SNP marker P297 was recognised in 2008 as ancestral to the significant subclades M73 and M269, combining them into one cluster.[7] This had been given the phylogenetic name R1b1a1a (and, previously, R1b1a).
A majority of Eurasian R1b falls within this subclade, representing a very large modern population. Although P297 itself has not yet been much tested for, the same population has been relatively well studied in terms of other markers. Therefore, the branching within this clade can be explained in relatively high detail below.
R1b1a1a (R-M73)
Malyarchuk et al. (2011) found R-M73 in 13.2% (5/38) of Shors, 11.4% (5/44) of Teleuts, 3.3% (2/60) of Kalmyks, 3.1% (2/64) of Khakassians, 1.9% (2/108) of Tuvinians, and 1.1% (1/89) of Altaians.[45] The Kalmyks, Tuvinians, and Altaian belong to a Y-STR cluster marked by DYS390=19, DYS389=14-16 (or 14–15 in the case of the Altaian individual), and DYS385=13-13.
Dulik et al. (2012) found R-M73 in 35.3% (6/17) of a sample of the Kumandin of the Altai Republic in Russia.[46] Three of these six Kumandins share an identical 15-loci Y-STR haplotype, and another two differ only at the DYS458 locus, having DYS458=18 instead of DYS458=17. This pair of Kumandin R-M73 haplotypes resembles the haplotypes of two Kalmyks, two Tuvinians, and one Altaian whose Y-DNA has been analyzed by Malyarchuk et al. (2011). The remaining R-M73 Kumandin has a Y-STR haplotype that is starkly different from the haplotypes of the other R-M73 Kumandins, resembling instead the haplotypes of five Shors, five Teleuts, and two Khakassians.[45]
While early research into R-M73 claimed that it was significantly represented among the Hazara of Afghanistan and the Bashkirs of the Ural Mountains, this has apparently been overturned. For example, supporting material from a 2010 study by Behar et al. suggested that Sengupta et al. (2006) might have misidentified Hazara individuals, who instead belonged to "PQR2" as opposed to "R(xR1a)."[47][40][48] However, the assignment of these Hazaras' Y-DNA to the "PQR2" category by Behar et al. (2010) is probably ascribable to the habit that was popular for a while of labeling R-M269 as "R1b" or "R(xR1a)," with any members of R-M343 (xM269) being placed in a polyphyletic, catch-all "R*" or "P" category. Myres et al. (2011), Di Cristofaro et al. (2013), and Lippold et al. (2014) all agree that the Y-DNA of 32% (8/25) of the HGDP sample of Pakistani Hazara should belong to haplogroup R-M478/M73.[40][49][50] Likewise, most Bashkir males have been found to belong to U-152 (R1b1a1a2a1a2b) and some, mostly from southeastern Bashkortostan, belonged to Haplogroup Q-M25 (Q1a1b) rather than R1b; contra this, Myres et al. (2011) found a high frequency of R-M73 among their sample of Bashkirs from southeast Bashkortostan (77/329 = 23.4% R1b-M73), in agreement with the earlier study of Bashkirs.[40] Besides the high frequency of R-M73 in southeastern Bashkirs, Myres et al. also reported finding R-M73 in the following samples: 10.3% (14/136) of Balkars from the northwest Caucasus, 9.4% (8/85) of the HGDP samples from northern Pakistan (these are the aforementioned Pakistani Hazaras), 5.8% (4/69) of Karachays from the northwest Caucasus, 2.6% (1/39) of Tatars from Bashkortostan, 1.9% (1/54) of Bashkirs from southwest Bashkortostan, 1.5% (1/67) of Megrels from the south Caucasus, 1.4% (1/70) of Bashkirs from north Bashkortostan, 1.3% (1/80) of Tatars from Kazan, 1.1% (1/89) of a sample from Cappadocia, Turkey, 0.7% (1/141) of Kabardians from the northwest Caucasus, 0.6% (3/522) of a pool of samples from Turkey, and 0.38% (1/263) of Russians from Central Russia.[40]
Besides the aforementioned Pakistani Hazaras, Di Cristofaro et al. (2013) found R-M478/M73 in 11.1% (2/18) of Mongols from central Mongolia, 5.0% (1/20) of Kyrgyz from southwest Kyrgyzstan, 4.3% (1/23) of Mongols from southeast Mongolia, 4.3% (4/94) of Uzbeks from Jawzjan, Afghanistan, 3.7% (1/27) of Iranians from Gilan, 2.5% (1/40) of Kyrgyz from central Kyrgyzstan, 2.1% (2/97) of Mongols from northwest Mongolia, and 1.4% (1/74) of Turkmens from Jawzjan, Afghanistan.[49] The Mongols as well as the individual from southwest Kyrgyzstan, the individual from Gilan, and one of the Uzbeks from Jawzjan belong to the same Y-STR haplotype cluster as five of six Kumandin members of R-M73 studied by Dulik et al. (2012). This cluster's most distinctive Y-STR value is DYS390=19.[40]
Karafet et al. (2018) found R-M73 in 37.5% (15/40) of a sample of Teleuts from Bekovo, Kemerovo oblast, 4.5% (3/66) of a sample of Uyghurs from Xinjiang Uyghur Autonomous Region, 3.4% (1/29) of a sample of Kazakhs from Kazakhstan, 2.3% (3/129) of a sample of Selkups, 2.3% (1/44) of a sample of Turkmens from Turkmenistan, and 0.7% (1/136) of a sample of Iranians from Iran.[51] Four of these individuals (one of the Teleuts, one of the Uyghurs, the Kazakh, and the Iranian) appear to belong to the aforementioned cluster marked by DYS390=19 (the Kumandin-Mongol R-M73 cluster); the Teleut and the Uyghur also share the modal values at the DYS385 and the DYS389 loci. The Iranian differs from the modal for this cluster by having 13-16 (or 13–29) at DYS389 instead of 14-16 (or 14–30). The Kazakh differs from the modal by having 13–14 at DYS385 instead of 13-13. The other fourteen Teleuts and the three Selkups appear to belong to the Teleut-Shor-Khakassian R-M73 cluster from the data set of Malyarchuk et al. (2011); this cluster has the modal values of DYS390=22 (but 21 in the case of two Teleuts and one Khakassian), DYS385=13-16, and DYS389=13-17 (or 13–30, but 14–31 in the case of one Selkup).
A Kazakhstani paper published in 2017 found haplogroup R1b-M478 Y-DNA in 3.17% (41/1294) of a sample of Kazakhs from Kazakhstan, with this haplogroup being observed with greater than average frequency among members of the Qypshaq (12/29 = 41.4%), Ysty (6/57 = 10.5%), Qongyrat (8/95 = 8.4%), Oshaqty (2/29 = 6.9%), Kerey (1/28 = 3.6%), and Jetyru (3/86 = 3.5%) tribes.[52] A Chinese paper published in 2018 found haplogroup R1b-M478 Y-DNA in 9.2% (7/76) of a sample of Dolan Uyghurs from Horiqol township, Awat County, Xinjiang.[53]
R1b1a1b (R-M269)
R-M269, or R1b1a1b (as of 2018) amongst other names,[54] is now the most common Y-DNA lineage in European males. It is carried by an estimated 110 million males in Europe.[55]
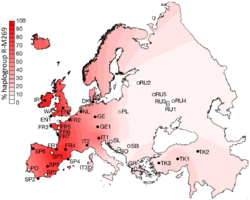
R-M269 has received significant scientific and popular interest due to its possible connection to the Indo-European expansion in Europe. Specifically the R-Z2103 subclade has been found to be prevalent in ancient DNA associated with the Yamna culture.[2] All seven individuals in one were determined to belong to the R1b-M269 subclade.[2]
Older research, published before researchers could study the DNA of ancient remains, proposed that R-M269 likely originated in Western Asia and was present in Europe by the Neolithic period.[35][40][56][57] But results based on actual ancient DNA noticed that there was a dearth of R-M269 in Europe before the Bronze Age,[2] and the distribution of subclades within Europe is substantially due to the various migrations of the Bronze and Iron Age. Likewise, the oldest samples classified as belonging to R-M269, have been found in Eastern Europe and Pontic-Caspian steppe, not Western Asia. Western European populations are divided between the R-P312/S116 and R-U106/S21 subclades of R-M412 (R-L51).
Distribution of R-M269 in Europe increases in frequency from east to west. It peaks at the national level in Wales at a rate of 92%, at 82% in Ireland, 70% in Scotland, 68% in Spain , 60% in France (76% in Normandy), about 60% in Portugal,[40] 50% in Germany , 50% in the Netherlands, 47% in Italy,[58] 45% in Eastern England and 42% in Iceland. R-M269 reaches levels as high as 95% in parts of Ireland. It has also been found at lower frequencies throughout central Eurasia,[59] but with relatively high frequency among the Bashkirs of the Perm region (84.0%).[60] This marker is present in China and India at frequencies of less than one percent. In North Africa and adjoining islands, while R-V88 (R1b1b) is more strongly represented, R-M269 appears to have been present since antiquity. R-M269 has been found, for instance, at a rate of ~44% among remains dating from the 11th to 13th centuries at Punta Azul, in the Canary Islands. These remains have been linked to the Bimbache (or Bimape), a subgroup of the Guanche.[61] In living males, it peaks in parts of North Africa, especially Algeria, at a rate of 10%.[62] In Sub-Saharan Africa, R-M269 appears to peak in Namibia, at a rate of 8% among Herero males.[63] In western Asia, R-M269 has been reported in 40% of Armenian males and over 35% in Turkmen males.[64][65] (The table below lists in more detail the frequencies of M269 in regions in Asia, Europe, and Africa.)
Apart from basal R-M269* which has not diverged, there are (as of 2017) two primary branches of R-M269:
- R-L23 (R1b1a1b1; L23/PF6534/S141) and
- R-PF7558 (R1b1a1b2; PF7558/PF7562.)
R-L23 (Z2105/Z2103; a.k.a. R1b1a1b1) has been reported among the peoples of the Idel-Ural (by Trofimova et al. 2015): 21 out of 58 (36.2%) of Burzyansky District Bashkirs, 11 out of 52 (21.2%) of Udmurts, 4 out of 50 (8%) of Komi, 4 out of 59 (6.8%) of Mordvins, 2 out of 53 (3.8%) of Besermyan and 1 out of 43 (2.3%) of Chuvash were R1b-L23.[66]
Subclades within the paragroup R-M269(xL23) – that is, R-M269* and/or R-PF7558 – appear to be found at their highest frequency in the central Balkans, especially Kosovo with 7.9%, North Macedonia 5.1% and Serbia 4.4%.[40] Unlike most other areas with significant percentages of R-L23, Kosovo, Poland and the Bashkirs of south-east Bashkortostan are notable in having a high percentage of R-L23 (xM412) – at rates of 11.4% (Kosovo), 2.4% (Poland) and 2.4% south-east Bashkortostan.[40] (This Bashkir population is also notable for its high level of R-M73 (R1b1a1a1), at 23.4%.[40]) Five individuals out of 110 tested in the Ararat Valley of Armenia belonged to R-M269(xL23) and 36 to R-L23*, with none belonging to known subclades of L23.[67]
In 2009, DNA extracted from the femur bones of 6 skeletons in an early-medieval burial place in Ergolding (Bavaria, Germany) dated to around AD 670 yielded the following results: 4 were found to be haplogroup R1b with the closest matches in modern populations of Germany, Ireland and the USA while 2 were in Haplogroup G2a.[68]
The following gives a summary of most of the studies which specifically tested for M269, showing its distribution (as a percentage of total population) in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.
The phylogeny of R-M269 according to ISOGG 2017:
| M269/PF6517 |
| |||||||||||||||||||||||||||||||||||||||||||||
R1b1b (R-V88)
R1b1b (PF6279/V88; previously R1b1a2) is defined by the presence of SNP marker V88, the discovery of which was announced in 2010 by Cruciani et al.[43] Apart from individuals in southern Europe and Western Asia, the majority of R-V88 was found in the Sahel, especially among populations speaking Afroasiatic languages of the Chadic branch.
Studies in 2005–08 reported "R1b*" at high levels in Jordan, Egypt and Sudan.[69][63][70][note 1] Subsequent research by Myres et al. (2011) indicates that the samples concerned most likely belong to the subclade R-V88, which is now concentrated in Sub-Saharan Africa. According to Myres et al. (2011), this may be explained by a back-migration from Asia into Africa by R1b-carrying people.[40][note 2] Gonzales et al. (2013), using more advanced techniques, indicate that it is equally probable that V88 originated in Central Africa and spread northward towards Asia. The patterns of diversity in African R1b-V88 did not seem to fit with a movement of Chadic-speaking people from the North, across the Sahara to West-Central Africa, but was compatible with the reverse. An origin of V88 lineages in Central Africa, followed by a migration to North Africa.[71] However, Shriner, D., & Rotimi, C. N. (2018), associate the introduction of R1b into Chad, with the more recent movements of Baggara Arabs.[72]
D'Atanasio et al. (2018) propose that R1b-V88 originated in Europe about 12 000 years ago and crossed to North Africa by about 8000 years ago; it may formerly have been common in southern Europe, where it has since been replaced by waves of other haplogroups, leaving remnant subclades almost exclusively in Sardinia. It first radiated within Africa likely between 8000 and 7000 years ago – at the same time as trans-Saharan expansions within the unrelated haplogroups E-M2 and A-M13 – possibly due to population growth allowed by humid conditions and the adoption of livestock herding in the Sahara. R1b-V1589, the main subclade within R1b-V88, underwent a further expansion around 5500 years ago, likely in the Lake Chad Basin region, from which some lines recrossed the Sahara to North Africa.[73]
Marcus et al. (2020) provide strong evidence for this proposed model of North to South trans-Saharan movement: The earliest basal R1b-V88 haplogroups are found in several Eastern European Hunter Gatherers close to 10 000 years ago. The haplogroup then seemingly further spread with the Neolithic Cardial Ware expansion, which established agriculture in the Western Mediterranean around 7500 BP: R1b-V88 haplogroups were identified in ancient Neolithic individuals in central Italy, Iberia and, at a particularly high frequency, in Sardinia.[74] A part of the branch leading to present-day African haplogroups (V2197) is already derived in some of these ancient Neolithic European individuals, providing further support for a North to South trans-Saharan movement.
| Region | Population | Country | Language | Sample size |
Total % |
R1b1b (R-V88) |
R1b1a1b (R-M269) |
R1b1b* (R-V88*) |
R1b1b2a2a1 (R-V69) |
|---|---|---|---|---|---|---|---|---|---|
| N Africa | Composite | Morocco | AA | 338 | 0.0% | 0.3% | 0.6% | 0.3% | 0.0% |
| N Africa | Mozabite Berbers | Algeria | AA/Berber | 67 | 3.0% | 3.0% | 0.0% | 3.0% | 0.0% |
| N Africa | Northern Egyptians | Egypt | AA/Semitic | 49 | 6.1% | 4.1% | 2.0% | 4.1% | 0.0% |
| N Africa | Berbers from Siwa | Egypt | AA/Berber | 93 | 28.0% | 26.9% | 1.1% | 23.7% | 3.2% |
| N Africa | Baharia | Egypt | AA/Semitic | 41 | 7.3% | 4.9% | 2.4% | 0.0% | 4.9% |
| N Africa | Gurna Oasis | Egypt | AA/Semitic | 34 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| N Africa | Southern Egyptians | Egypt | AA/Semitic | 69 | 5.8% | 5.8% | 0.0% | 2.9% | 2.9% |
| C Africa | Songhai | Niger | NS/Songhai | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Fulbe | Niger | NC/Atlantic | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Tuareg | Niger | AA/Berber | 22 | 4.5% | 4.5% | 0.0% | 4.5% | 0.0% |
| C Africa | Ngambai | Chad | NS/Sudanic | 11 | 9.1% | 9.1% | 0.0% | 9.1% | 0.0% |
| C Africa | Hausa | Nigeria (North) | AA/Chadic | 10 | 20.0% | 20.0% | 0.0% | 20.0% | 0.0% |
| C Africa | Fulbe | Nigeria (North) | NC/Atlantic | 32 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Yoruba | Nigeria (South) | NC/Defoid | 21 | 4.8% | 4.8% | 0.0% | 4.8% | 0.0% |
| C Africa | Ouldeme | Cameroon (Nth) | AA/Chadic | 22 | 95.5% | 95.5% | 0.0% | 95.5% | 0.0% |
| C Africa | Mada | Cameroon (Nth) | AA/Chadic | 17 | 82.4% | 82.4% | 0.0% | 76.5% | 5.9% |
| C Africa | Mafa | Cameroon (Nth) | AA/Chadic | 8 | 87.5% | 87.5% | 0.0% | 25.0% | 62.5% |
| C Africa | Guiziga | Cameroon (Nth) | AA/Chadic | 9 | 77.8% | 77.8% | 0.0% | 22.2% | 55.6% |
| C Africa | Daba | Cameroon (Nth) | AA/Chadic | 19 | 42.1% | 42.1% | 0.0% | 36.8% | 5.3% |
| C Africa | Guidar | Cameroon (Nth) | AA/Chadic | 9 | 66.7% | 66.7% | 0.0% | 22.2% | 44.4% |
| C Africa | Massa | Cameroon (Nth) | AA/Chadic | 7 | 28.6% | 28.6% | 0.0% | 14.3% | 14.3% |
| C Africa | Other Chadic | Cameroon (Nth) | AA/Chadic | 4 | 75.0% | 75.0% | 0.0% | 25.0% | 50.0% |
| C Africa | Shuwa Arabs | Cameroon (Nth) | AA/Semitic | 5 | 40.0% | 40.0% | 0.0% | 40.0% | 0.0% |
| C Africa | Kanuri | Cameroon (Nth) | NS/Saharan | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Fulbe | Cameroon (Nth) | NC/Atlantic | 18 | 11.1% | 11.1% | 0.0% | 5.6% | 5.6% |
| C Africa | Moundang | Cameroon (Nth) | NC/Adamawa | 21 | 66.7% | 66.7% | 0.0% | 14.3% | 52.4% |
| C Africa | Fali | Cameroon (Nth) | NC/Adamawa | 48 | 20.8% | 20.8% | 0.0% | 10.4% | 10.4% |
| C Africa | Tali | Cameroon (Nth) | NC/Adamawa | 22 | 9.1% | 9.1% | 0.0% | 4.5% | 4.5% |
| C Africa | Mboum | Cameroon (Nth) | NC/Adamawa | 9 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Composite | Cameroon (Sth) | NC/Bantu | 90 | 0.0% | 1.1% | 0.0% | 1.1% | 0.0% |
| C Africa | Biaka Pygmies | CAR | NC/Bantu | 33 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| W Africa | Composite | — | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| E Africa | Composite | — | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| S Africa | Composite | — | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Total | 1822 | ||||||||
| V88 |
| ||||||||||||
Two branches of R-V88, R-M18 and R-V35, are found almost exclusively on the island of Sardinia.
As can be seen in the above data table, R-V88 is found in northern Cameroon in west central Africa at a very high frequency, where it is considered to be caused by a pre-Islamic movement of people from Eurasia.[63][75]
R1b1b1 (R-M18)
R1b1b1 is a sub-clade of R-V88, which is defined by the presence of SNP marker M18.[7] It has been found only at low frequencies in samples from Sardinia[59][76] and Lebanon.[77]
R1b2 (R-PH155)
R1b2 is extremely rare and defined by the presence of PH155.[35] Living males carrying subclades of R-PH155 have been found in Bahrain, Bhutan, Ladakh, Tajikistan, Turkey, Xinjiang, and Yunnan. ISOGG (2022) cites two primary branches: R-M335 (R1b2a) and R-PH200 (R1b2b).
The defining SNP of R1b2a, M335, was first documented in 2004, when an example was discovered in Turkey, though it was classified at that time as R1b4.[33] Other examples of R-M335 have been reported in a sample of Hui from Yunnan, China[78] and in a sample of people from Ladakh, India.[79]
In popular culture
- Bryan Sykes, in his 2006 book Blood of the Isles, gives the members – and the notional founding patriarch – of R1b the name "Oisín".
- Stephen Oppenheimer, in his 2007 book Origins of the British, gives the R1b patriarch the Basque name "Ruisko" in honour of what Oppenheimer believed to be the Iberian origin of R1b.
- A filmmaker named Artem Lukichev created (circa 2009) a 14-minute animated film based on a Bashkir epic from the Ural Mountains, relating the epic to the emergence and geographical expansion of R1a and R1b.[80]
- DNA tests that assisted in the identification of Czar Nicholas II of Russia found that he belonged to R1b.[81] This may suggest that the later Czars of the House of Romanov, descended in the male line from the House of Holstein-Gottorp (which originated in Schleswig-Holstein) are also members of R1b.
Health
Studies have shown that haplogroup R1b could have a protective effect on the immune system.[82] However, later studies have confirmed that the Y-chromosome has a very limited effect on coronary artery disease (CAD), for example, and that the previously purported link between Y-chromosome haplogroups and health is far from established scientifically.[83]
See also
- Atlantic modal haplotype
- Genealogical DNA test
- Y-DNA haplogroups in populations of Europe
Notes
- ↑ Flores et al. (2005) found that 20 out of all 146 men tested (13.7%) – including 20 out of 45 men tested from the Dead Sea area of Jordan – were positive for M173 (R1), and negative for both the R1a markers SRY10831.2 and M17, as well as P25 (which was later discovered to be an unreliable marker for R1b1). Myres et al. (2011)[40] indicates that they are mostly R-V88 (later known as R1b1b). Wood et al. (2005) also reported two Egyptian cases of R1* (R-M173*) that were negative for SRY10831 (R1a1) and the unreliable R1b1 marker P25, out of a sample of 1,122 males from African countries, including 92 from Egypt. Hassan et al. (2008) found an equally surprising 14 out of 26 (54%) of Sudanese Fula people who were M173+ and P25-.
- ↑ Myres et al. (2011): "The detection of V88 in Iran, Palestine and especially the Dead Sea, Jordan (Supplementary Table S4) provides an insight into the back to Africa migration route.
References
- ↑ 1.0 1.1 1.2 "The genetic history of Ice Age Europe". Nature 534 (7606): 200–5. June 2016. doi:10.1038/nature17993. PMID 27135931. Bibcode: 2016Natur.534..200F.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Haak et al. 2015.
- ↑ 3.0 3.1 "Population genomics of Bronze Age Eurasia". Nature 522 (7555): 167–72. June 2015. doi:10.1038/nature14507. PMID 26062507. Bibcode: 2015Natur.522..167A. https://depot.ceon.pl/handle/123456789/13155.
- ↑ 4.0 4.1 "Eight thousand years of natural selection in Europe". bioRxiv: 016477. 2015. doi:10.1101/016477.
- ↑ "Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome". Proceedings of the National Academy of Sciences of the United States of America 113 (2): 368–73. January 2016. doi:10.1073/pnas.1518445113. PMID 26712024. Bibcode: 2016PNAS..113..368C.
- ↑ "The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods". PLOS Genetics 13 (7): e1006852. July 2017. doi:10.1371/journal.pgen.1006852. PMID 28749934.
- ↑ 7.0 7.1 7.2 "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. May 2008. doi:10.1101/gr.7172008. PMID 18385274.
- ↑ Mathieson 2018, Supplementary Table 1, Row 467.
- ↑ Mathieson 2018, Supplementary Table 1, Rows 251-272.
- ↑ 10.0 10.1 Jones 2017.
- ↑ Mathieson 2018, Supplementary Table 1, Rows 205-245.
- ↑ Mathieson 2018, Supplementary Table 1.
- ↑ Mathieson 2018, pp. 2-3.
- ↑ González-Fortes et al. 2017, pp. 8-9.
- ↑ González-Fortes et al. 2017, p. 4, Table 1, OC1_Meso.
- ↑ Sánchez-Quinto et al. 2019, Extended Dataset 1.3, OC1.
- ↑ Mathieson 2018, Supplementary Table 1, Row 298, I4666.
- ↑ Mathieson 2018, Supplementary Table 1, Row 153, I0124.
- ↑ Mathieson 2018, Supplementary Table 1, Row 375, I0122.
- ↑ Haak et al. 2015, Supplementary Information, pp. 26-27.
- ↑ Haak et al. 2015, Extended Data Table 2, I0410.
- ↑ Haak et al. 2015, Supplementary Information, pp. 44-45.
- ↑ Mathieson 2018, Supplementary Table 1, I0410.
- ↑ Mathieson 2018, Supplementary Table 1, Row 73, I2430.
- ↑ Marcus et al. 2020, Supplementary Data 1, A Master Table, Row 25, MA89.
- ↑ Mathieson 2018, Supplementary Table 1, Row 128, I0559.
- ↑ "The first horse herders and the impact of early Bronze Age steppe expansions into Asia". Science 360 (6396): eaar7711. June 2018. doi:10.1126/science.aar7711. PMID 29743352.
- ↑ Haak et al. 2015, p. 5.
- ↑ "Improved phylogenetic resolution and rapid diversification of Y-chromosome haplogroup K-M526 in Southeast Asia". European Journal of Human Genetics 23 (3): 369–73. March 2015. doi:10.1038/ejhg.2014.106. PMID 24896152.
- ↑ "Ancient DNA reveals male diffusion through the Neolithic Mediterranean route". Proceedings of the National Academy of Sciences of the United States of America 108 (24): 9788–91. June 2011. doi:10.1073/pnas.1100723108. PMID 21628562. Bibcode: 2011PNAS..108.9788L.
- ↑ "Ancient DNA from European early neolithic farmers reveals their near eastern affinities". PLOS Biology 8 (11): e1000536. November 2010. doi:10.1371/journal.pbio.1000536. PMID 21085689.
- ↑ "The Beaker phenomenon and the genomic transformation of northwest Europe". Nature 555 (7695): 190–196. March 2018. doi:10.1038/nature25738. PMID 29466337. Bibcode: 2018Natur.555..190O.
- ↑ 33.0 33.1 33.2 "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. January 2004. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- ↑ 34.0 34.1 "ISOGG 2016 Y-DNA Haplogroup R". https://isogg.org/tree/2016/ISOGG_HapgrpR16.html.
- ↑ 35.0 35.1 35.2 "ISOGG 2017 Y-DNA Haplogroup R". https://isogg.org/tree/ISOGG_HapgrpR.html.
- ↑ YFull YTree v10.00.00
- ↑ Phylogenetic tree of haplogroup R-V1636 at 23mofang
- ↑ Haplogroup R on the Y-DNA Haplotree at FTDNA
- ↑ "R-V1636 YTree". https://www.yfull.com/tree/R-V1636/.
- ↑ 40.00 40.01 40.02 40.03 40.04 40.05 40.06 40.07 40.08 40.09 40.10 40.11 40.12 "A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe". European Journal of Human Genetics 19 (1): 95–101. January 2011. doi:10.1038/ejhg.2010.146. PMID 20736979.
- ↑ "Ancient migratory events in the Middle East: new clues from the Y-chromosome variation of modern Iranians". PLOS ONE 7 (7): e41252. July 18, 2012. doi:10.1371/journal.pone.0041252. PMID 22815981. Bibcode: 2012PLoSO...741252G.
- ↑ "The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion". Forensic Science International 159 (1): 14–20. May 2006. doi:10.1016/j.forsciint.2005.06.003. PMID 16026953. https://figshare.com/articles/journal_contribution/10076234.
- ↑ 43.0 43.1 43.2 "Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages". European Journal of Human Genetics 18 (7): 800–7. July 2010. doi:10.1038/ejhg.2009.231. PMID 20051990.
- ↑ Varzari A (2006). Population History of the Dniester-Carpathians: evidence from Alu insertion and Y-chromosome polymorphisms (Thesis). Ludwig-Maximilians-Universität München. doi:10.5282/edoc.5868.
- ↑ 45.0 45.1 "Ancient links between Siberians and Native Americans revealed by subtyping the Y chromosome haplogroup Q1a". Journal of Human Genetics 56 (8): 583–8. August 2011. doi:10.1038/jhg.2011.64. PMID 21677663.
- ↑ "Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between Native Americans and Indigenous Altaians". American Journal of Human Genetics 90 (2): 229–46. February 2012. doi:10.1016/j.ajhg.2011.12.014. PMID 22281367.
- ↑ "The genome-wide structure of the Jewish people". Nature 466 (7303): 238–42. July 2010. doi:10.1038/nature09103. PMID 20531471. Bibcode: 2010Natur.466..238B.
- ↑ "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". American Journal of Human Genetics 78 (2): 202–21. February 2006. doi:10.1086/499411. PMID 16400607.
- ↑ 49.0 49.1 "Afghan Hindu Kush: where Eurasian sub-continent gene flows converge". PLOS ONE 8 (10): e76748. 2013. doi:10.1371/journal.pone.0076748. PMID 24204668. Bibcode: 2013PLoSO...876748D.
- ↑ "Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences". Investigative Genetics 5: 13. 2014. doi:10.1186/2041-2223-5-13. PMID 25254093.
- ↑ "Siberian genetic diversity reveals complex origins of the Samoyedic-speaking populations". American Journal of Human Biology 30 (6): e23194. November 2018. doi:10.1002/ajhb.23194. PMID 30408262.
- ↑ "Распределение гаплогрупп И-хромосомы казахов Южно-Казахстанской, Жамбылской и Алматинской областей" (in ru). Reports of the National Academy of Sciences of the Republic of Kazakhstan 6 (316): 85–95. 2017. http://nblib.library.kz/elib/library.kz/jurnal/%D0%94%D0%BE%D0%BA%D0%BB%D0%B0%D0%B4_06_2017%20(2)/12-%20Biology%20E.E.Ashirbekov0617.pdf. Retrieved 2021-11-20.
- ↑ "A study of genetic diversity of three isolated populations in Xinjiang using Y-SNP.". Acta Anthropologica Sinica 37 (1): 146–56. 2018. doi:10.16359/j.cnki.cn11-1963/q.2017.0067.
- ↑ R1b1a1b (R-M269) was previously R1b1a1a2. From 2003 to 2005, what is now R1b1a1b was designated R1b3. From 2005 to 2008, it was R1b1c. From 2008 to 2011, it was R1b1b2. From 2011 to 2018, it was R1b1a1a2.
- ↑ 55.0 55.1 "A predominantly neolithic origin for European paternal lineages". PLOS Biology 8 (1): e1000285. January 2010. doi:10.1371/journal.pbio.1000285. PMID 20087410.
- ↑ "The peopling of Europe". Anthropological genetics: theory, methods and applications. Cambridge, UK: Cambridge University Press. 2007. p. 394. ISBN 978-0-521-54697-3.
- ↑ "Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152". Forensic Science International. Genetics 5 (3): e49-52. June 2011. doi:10.1016/j.fsigen.2010.07.006. PMID 20732840.
- ↑ Grugni, Viola; Raveane, Alessandro; Mattioli, Francesca; Battaglia, Vincenza; Sala, Cinzia; Toniolo, Daniela; Ferretti, Luca; Gardella, Rita et al. (February 2018). "Reconstructing the genetic history of Italians: new insights from a male (Y-chromosome) perspective". Annals of Human Biology 45 (1): 44–56. doi:10.1080/03014460.2017.1409801. ISSN 1464-5033. PMID 29382284. https://pubmed.ncbi.nlm.nih.gov/29382284/.
- ↑ 59.0 59.1 "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. November 2000. doi:10.1038/81685. PMID 11062480.
- ↑ Lobov AS (2009). Structure of the Gene Pool of Bashkir Subpopulations (PDF) (Ph.D. thesis) (in русский). Institute of Biochemistry and Genetics of the Ufa Scientific Center of the Russian Academy of Sciences. Archived from the original (PDF) on 2011-08-16.
- ↑ "Genetic studies on the prehispanic population buried in Punta Azul cave (El Hierro, Canary Islands)". Journal of Archaeological Science 78: 20–28. 2017. doi:10.1016/j.jas.2016.11.004. Bibcode: 2017JArSc..78...20O.
- ↑ "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample". International Journal of Legal Medicine 122 (3): 251–5. May 2008. doi:10.1007/s00414-007-0203-5. PMID 17909833.
- ↑ 63.0 63.1 63.2 "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes". European Journal of Human Genetics 13 (7): 867–76. July 2005. doi:10.1038/sj.ejhg.5201408. PMID 15856073.
- ↑ "Genetic Testing of Language Replacement Hypothesis in Southwest Asia". Iran and the Caucasus 10 (2): 191–208. 2006. doi:10.1163/157338406780345899.
- ↑ Maciamo Hay. Haplogrouop R1b (Y-DNA). https://www.eupedia.com/europe/Haplogroup_R1b_Y-DNA.shtml.
- ↑ Vadimovna TN (February 2015). Изменчивость Митохондриальной ДНК и Y-Хромосомы в Популяциях Волго-Уральского Региона [Mitochondrial DNA variation and the Y-chromosome in the population of the Volga-Ural Region] (PDF) (Biological Sciences thesis) (in Russian). Archived from the original (PDF) on 2017-04-02.CS1 maint: unrecognized language (link)
- ↑ "Neolithic patrilineal signals indicate that the Armenian plateau was repopulated by agriculturalists". European Journal of Human Genetics 20 (3): 313–20. March 2012. doi:10.1038/ejhg.2011.192. PMID 22085901.
- ↑ "Kinship and Y-chromosome analysis of 7th century human remains: novel DNA extraction and typing procedure for ancient material". Croatian Medical Journal. 3 50 (3): 286–95. June 2009. doi:10.3325/cmj.2009.50.286. PMID 19480023.
- ↑ "Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan". Journal of Human Genetics 50 (9): 435–441. 2005. doi:10.1007/s10038-005-0274-4. PMID 16142507.
- ↑ "Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history". American Journal of Physical Anthropology 137 (3): 316–323. November 2008. doi:10.1002/ajpa.20876. PMID 18618658. "13/32".
- ↑ "The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88". European Journal of Human Genetics 21 (3): 324–331. March 2013. doi:10.1038/ejhg.2012.167. PMID 22892526.
- ↑ Shriner, Daniel; Rotimi, Charles N. (December 2018). "Genetic History of Chad". American Journal of Physical Anthropology 167 (4): 804–812. doi:10.1002/ajpa.23711. ISSN 0002-9483. PMID 30259956.
- ↑ "The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages". Genome Biology 19 (1): 20. February 2018. doi:10.1186/s13059-018-1393-5. PMID 29433568.
- ↑ "Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia". Nature Communications 11 (1): 939. February 2020. doi:10.1038/s41467-020-14523-6. PMID 32094358. Bibcode: 2020NatCo..11..939M.
- ↑ "A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes". American Journal of Human Genetics 70 (5): 1197–214. May 2002. doi:10.1086/340257. PMID 11910562., pp. 13–14
- ↑ "Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse Sardinian population: inference for association scans". PLOS ONE 3 (1): e1430. January 2008. doi:10.1371/journal.pone.0001430. PMID 18183308. Bibcode: 2008PLoSO...3.1430C.
- ↑ "Y-chromosomal diversity in Lebanon is structured by recent historical events". American Journal of Human Genetics 82 (4): 873–882. April 2008. doi:10.1016/j.ajhg.2008.01.020. PMID 18374297.
- ↑ "Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route". Molecular Biology and Evolution 28 (1): 717–27. January 2011. doi:10.1093/molbev/msq247. PMID 20837606.
- ↑ "Ladakh, India: the land of high passes and genetic heterogeneity reveals a confluence of migrations". European Journal of Human Genetics 24 (3): 442–9. March 2016. doi:10.1038/ejhg.2015.80. PMID 25966630.
- ↑ "About R1a and R1b from Ural epic story. Artem Lukichev (c)". https://www.youtube.com/watch?v=wMBuo7JHesg.
- ↑ "Mystery solved: the identification of the two missing Romanov children using DNA analysis". PLOS ONE 4 (3): e4838. 2009. doi:10.1371/journal.pone.0004838. PMID 19277206. Bibcode: 2009PLoSO...4.4838C.
- ↑ "The Y chromosome: a blueprint for men's health?". European Journal of Human Genetics 25 (11): 1181–1188. November 2017. doi:10.1038/ejhg.2017.128. PMID 28853720.
- ↑ "Limited Effect of Y Chromosome Variation on Coronary Artery Disease and Mortality in UK Biobank—Brief Report". Arteriosclerosis, Thrombosis, and Vascular Biology 42 (9): 1198–1206. July 2022. doi:10.1161/ATVBAHA.122.317664. PMID 35861954.
Bibliography
- "The Neolithic Transition in the Baltic Was Not Driven by Admixture with Early European Farmers". Current Biology 27 (4): 576–582. February 2017. doi:10.1016/j.cub.2016.12.060. PMID 28162894.
- "Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin". Current Biology 27 (12): 1801–1810.e10. June 2017. doi:10.1016/j.cub.2017.05.023. PMID 28552360.
- "Massive migration from the steppe was a source for Indo-European languages in Europe". Nature 522 (7555): 207–211. June 2015. doi:10.1038/nature14317. PMID 25731166. Bibcode: 2015Natur.522..207H.
- "New genetic evidence of affinities and discontinuities between bronze age Siberian populations". American Journal of Physical Anthropology 167 (1): 97–107. September 2018. doi:10.1002/ajpa.23607. PMID 29900529.
- "Parallel palaeogenomic transects reveal complex genetic history of early European farmers". Nature 551 (7680): 368–372. November 2017. doi:10.1038/nature24476. PMID 29144465. Bibcode: 2017Natur.551..368L.
- "Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia". Nature Communications 11 (1): 939. February 2020. doi:10.1038/s41467-020-14523-6. PMID 32094358. Bibcode: 2020NatCo..11..939M.
- "The genomic history of southeastern Europe". Nature 555 (7695): 197–203. March 2018. doi:10.1038/nature25778. PMID 29466330. Bibcode: 2018Natur.555..197M.
- "The formation of human populations in South and Central Asia". Science 365 (6457): eaat7487. September 2019. doi:10.1126/science.aat7487. PMID 31488661.
- "The Beaker phenomenon and the genomic transformation of northwest Europe". Nature 555 (7695): 190–196. March 2018. doi:10.1038/nature25738. PMID 29466337. Bibcode: 2018Natur.555..190O.
- Haak, Wolfgang; Lazaridis, Iosif; Patterson, Nick; Rohland, Nadin; Mallick, Swapan; Llamas, Bastien; Brandt, Guido; Nordenfelt, Susanne et al. (2015). "Massive migration from the steppe is a source for Indo-European languages in Europe". Nature 522 (7555): 207–211. doi:10.1038/NATURE14317. PMID 25731166. Bibcode: 2015Natur.522..207H.
- "Megalithic tombs in western and northern Neolithic Europe were linked to a kindred society". Proceedings of the National Academy of Sciences of the United States of America 116 (19): 9469–9474. May 2019. doi:10.1073/pnas.1818037116. PMID 30988179. Bibcode: 2019PNAS..116.9469S.
- "The genetic history of Ice Age Europe". Nature 534 (7606): 200–5. June 2016. doi:10.1038/nature17993. PMID 27135931. Bibcode: 2016Natur.534..200F.
 |
