Biology:Lipid bilayer characterization
Lipid bilayer characterization is the use of various optical, chemical and physical probing methods to study the properties of lipid bilayers. Many of these techniques are elaborate and require expensive equipment because the fundamental nature of the lipid bilayer makes it a very difficult structure to study. An individual bilayer, since it is only a few nanometers thick, is invisible in traditional light microscopy. The bilayer is also a relatively fragile structure since it is held together entirely by non-covalent bonds and is irreversibly destroyed if removed from water. In spite of these limitations dozens of techniques have been developed over the last seventy years to allow investigations of the structure and function of bilayers. The first general approach was to utilize non-destructive in situ measurements such as x-ray diffraction and electrical resistance which measured bilayer properties but did not actually image the bilayer. Later, protocols were developed to modify the bilayer and allow its direct visualization at first in the electron microscope and, more recently, with fluorescence microscopy. Over the past two decades, a new generation of characterization tools including AFM has allowed the direct probing and imaging of membranes in situ with little to no chemical or physical modification. More recently, dual polarisation interferometry has been used to measure the optical birefringence of lipid bilayers to characterise order and disruption associated with interactions or environmental effects.
Fluorescence Microscopy
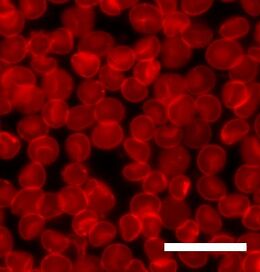
Fluorescence microscopy is a technique whereby certain molecules can be excited with one wavelength of light and will emit another longer wavelength of light. Because each fluorescent molecule has a unique spectrum of absorption and emission, the location of particular types of molecules can be determined. Natural lipids do not fluoresce, so it is always necessary to include a dye molecule in order to study lipid bilayers with fluorescence microscopy. To some extent, the addition of the dye molecule always changes the system, and in some cases it can be difficult to say whether the observed effect is due to the lipids, the dye or, most commonly, some combination of the two. The dye is usually attached either to a lipid or a molecule that closely resembles a lipid, but since the dye domain is relatively large it can alter the behavior of this other molecule. This is a particularly contentious issue when studying the diffusion or phase separation of lipids, as both processes are very sensitive to the size and shape of the molecules involved.
This potential complication has been given an argument against the use of one of fluorescence recovery after photobleaching (FRAP) to determine bilayer diffusion coefficients. In a typical FRAP experiment a small (~30 µm diameter) area is photobleached by exposure to an intense light source. This area is then monitored over time as the “dead” dye molecules diffuse out and are replaced by intact dye molecules from the surrounding bilayer. By fitting this recovery curve it is possible to calculate the diffusion coefficient of the bilayer.[1][2] An argument against the use of this technique is that what is actually being studied is the diffusion of the dye, not the lipid.[3] While correct, this distinction is not always important, since the mobility of the dye is often dominated by the mobility of the bilayer.
In traditional fluorescence microscopy the resolution has been limited to approximately half the wavelength of the light used. Through the use of confocal microscopy and image processing this limit can be extended, but typically not much below 100 nanometers, which is much smaller than a typical cell but much larger than the thickness of a lipid bilayer. More recently, advanced microscopy methods have allowed much greater resolution under certain circumstances, even down to sub-nm. One of the first of these methods to be developed was Förster resonance energy transfer (FRET). In FRET, two dye molecules are chosen such that the emission spectrum of one overlaps the absorption spectrum of the other. This energy transfer is extremely distance dependent, so it is possible to tell with angstrom resolution how far apart the two dyes are. This can be used for instance to determine when two bilayers fuse and their components mix.[4] Another high resolution microscopy technique is fluorescence interference contrast microscopy (FLIC). This method requires that the sample be mounted on a precisely micromachined reflective surface. By studying the destructive interference patterns formed it is possible to individually resolve the two leaflets of a supported bilayer and determine the distribution of a fluorescent dye in each.[5]
Electrical

Electrical measurements are the most straightforward way to characterize one of the more important functions of a bilayer, namely its ability to segregate and prevent the flow of ions in solution. Accordingly, electrical characterization was one of the first tools used to study the properties of model systems such as black membranes. It was already known that the cell membrane was capable of supporting an ionic gradient and that this gradient is responsible for the ability of neurons to send signals via an action potential. Demonstrating that similar phenomena could be replicated in vitro was an important verification of the utility of model systems.[6]
Fundamentally, all electrical measurements of bilayers involve the placement of an electrode on either side of the membrane. By applying a bias across these electrodes and measuring the resulting current, it is possible to determine the resistance of the bilayer. This resistance is typically quite high for intact bilayers, often exceeding 100 GΩ since the hydrophobic core is impermeable to charged hydrated species. Because this resistance is so large, the presence of even a few nanometer-scale holes results in a dramatic increase in current and can be easily determined.[7] The sensitivity of this system is such that even the activity of single ion channels can be resolved.[8] In such DC measurements, it is necessary to use electrochemically active electrodes to provide the necessary positive charges on one side and negative charges on the other. The most common system is the silver/silver chloride electrode since this reaction is stable, reversible, involves a single electron transfer and can produce large currents.[9] In addition to simple DC current measurements it is also possible to perform AC electrical characterization to extract information about the capacitance and complex impedance of a bilayer. Because capacitance is inversely proportional to thickness and bilayers are very thin they typically have a very large capacitance, on the order of 2µF/cm2. Capacitance measurements are particularly useful when dealing with black lipid membranes, as they can be used to determine when the solvent/lipid plug thins down to a single bilayer.
Optical
Lipids are highly polar molecules which when self assembled into bilayers creates a highly birefringent layer[10] where the optical properties parallel are very different from those perpendicular. This effect, studied by dual polarisation interferometry has been used to measure dynamic reorganisation of the layer due to temperature, ionic strength, and molecular interactions with e.g. antimicrobial peptides.
Hydrated bilayers show rich vibrational dynamics and are good media for efficient vibrational energy transfer. Vibrational properties of lipid monolayers and bilayers has been investigated by ultrafast spectroscopic techniques[11] and recently developed computational methods.[12]
AFM
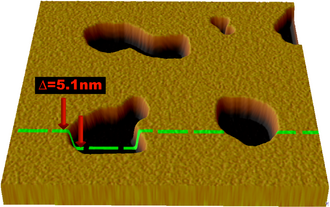
Atomic force microscopy (AFM) has been used in recent years to image and probe the physical properties of lipid bilayers. AFM is a promising technique because it has the potential to image with nanometer resolution at room temperature and even underwater, conditions necessary for natural bilayer behavior. These capabilities have allowed direct imaging of the subtle ripple phase transition in a supported bilayer.[13] Another AFM experiment performed in a tapping mode under aqueous buffer medium allowed (1) to determine the formation of transmembrane pores (holes) around nanoparticles of approximately 1.2 to 22 nm diameter via subtraction of AFM images from series recorded during the lipid bilayer formation and (2) to observe adsorption of single insulin molecules onto exposed nanoparticles.[14] Another advantage is that AFM does not require fluorescent or isotopic labeling of the lipids, as the probe tip interacts mechanically with the bilayer surface. Because of this, the same scan can reveal information about both the bilayer and any associated structures, even to the extent of resolving individual membrane proteins.[15] In addition to imaging, AFM can also probe the mechanical nature of small delicate structures such as lipid bilayers. One study demonstrated the possibility of measuring the elastic modulus of individual nano-scale membranes suspended over porous anodic alumina.[16]
Although AFM is a powerful and versatile tool for studying lipid bilayers, there are some practical limitations and difficulties. Because of the fragile nature of the bilayer, extremely low scanning forces (typically 50pN or less[13][17]) must be used to avoid damage. This consideration is particularly important when studying metastable systems such as vesicles adsorbed on a substrate, since the AFM tip can induce rupture and other structural changes.[18] Care must also be taken to choose an appropriate material and surface preparation for the AFM tip, as hydrophobic surfaces can interact strongly with lipids and disrupt the bilayer structure.[19]
Electron microscopy
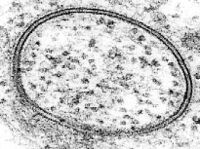
In electron microscopy a beam of focused electrons interacts with the sample rather than a beam of light as in traditional microscopy. Electrons have a much shorter wavelength than light so electron microscopy has much higher resolution than light microscopy, potentially down to the atomic scale. Because lipid bilayers are arranged on the molecular level, this higher resolution has been invaluable. In 1960, when the structure of the bilayer was still debated, it was electron microscopy that offered the first direct visualization of the two apposing leaflets.[20] In conjunction with rapid freezing techniques, electron microscopy has also been used to study the mechanisms of inter- and intracellular transport, for instance in demonstrating that exocytotic vesicles are the means of chemical release at synapses.[21] Often, electron microscopy is the only probe technique with sufficient resolution to determine complex nanometer-scale morphologies.
The limitations of electron microscopy in the study of lipid structures deal primarily with sample preparation. Most electron microscopes require the sample to be under vacuum, which is incompatible with hydration at room temperature. To surmount this problem, samples can be imaged under cryogenic conditions with the associated water frozen, or a metallic negative can be made from a frozen sample. It is also typically necessary to stain the bilayer with a heavy metal compound such as osmium tetroxide or uranyl acetate because the low atomic weight constituents of lipids (carbon, nitrogen, phosphorus, etc.) offer little contrast compared to water. If a Transmission electron microscope (TEM) is being used, it is also necessary to cut or polish the sample into a very thin (<1 micrometre) sheet, which can be difficult and time-consuming. Scanning Electron Microscopy (SEM) does not require this step, but cannot offer the same resolution as TEM. Both methods are surface-sensitive techniques and cannot reveal information about deeply buried structures.
Neutron and X-ray scattering
Both X-rays and high-energy neutrons are used to probe the structure and periodicity of biological structures including bilayers because they can be tuned to interact with matter at the relevant (angstrom-nm) length scales. Often, these two classes of experiment provide complementary information because each has different advantages and disadvantages. X-rays interact only weakly with water, so bulk samples can be probed with relatively easy sample preparation. This is one of the reasons that x-ray scattering was the technique first used to systematically study inter-bilayer spacing.[22] X-ray scattering can also yield information on the average spacing between individual lipid molecules, which has led to its use in characterizing phase transitions.[23] One limitation of x-ray techniques is that x-rays are relatively insensitive to light elements such as hydrogen. This effect is a consequence of the fact that x-rays interact with matter by scattering off of electron density which decreases with decreasing atomic number. In contrast, neutrons scatter off of nuclear density and nuclear magnetic fields so sensitivity does not decrease monotonically with z. This mechanism also provides strong isotopic contrast in some cases, notably between hydrogen and deuterium, allowing researchers to tune the experimental baseline by mixing water and deuterated water. Using reflectometry rather than scattering with neutrons or x-rays allow experimenters to probe supported bilayers or multilayer stacks. These measurements are more complicated to perform an analyze, but allow determination of cross sectional composition, including the location and concentration of water within the bilayer.[24] In the case of both neutron and x-ray scattering measurements, the information provided is an ensemble average of the system and is therefore subject to uncertainty based on thermal fluctuations in these highly mobile structures.[25]
References
- ↑ D. Axelrod, D. E. Koppel, J. Schlessinger, E. Elson and W. W. Webb."Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. ." Biophysical Journal. 16. (1976) 1055-69.
- ↑ D. M. Soumpasis."Theoretical analysis of fluorescence photobleaching recovery experiments." Biophysical Journal. 41. (1983) 95-7.
- ↑ W. L. Vaz and P. F. Almeida."Microscopic versus macroscopic diffusion in one-component fluid phase lipid bilayer membranes." Biophysical Journal. 60. (1991) 1553-1554.
- ↑ L. Guohua and R. C. Macdonald."Lipid bilayer vesicle fusion: Intermediates captured by high-speed microfluorescence spectroscopy." Biophysical Journal. 85. (2003) 1585-1599.
- ↑ J. M. Crane, V. Kiessling and L. K. Tamm."Measuring lipid asymmetry in planar supported bilayers by fluorescence interference contrast microscopy." Langmuir. 21. (2005) 1377-1388.
- ↑ P. Mueller, D. O. Rudin, H. I. Tien and W. C. Wescott."Reconstitution of cell membrane structure in vitro and its transformation into an excitable system." Nature. 194. (1962) 979-980.
- ↑ K. C. Melikov, V. A. Frolov, A. Shcherbakov, A. V. Samsonov, Y. A. Chizmadzhev and L. V. Chernomordik."Voltage-Induced Nonconductive Pre-Pores and Metastable Single Pores in Unmodified Planar Lipid Bilayer " Biophysical Journal. 80. (2001) 1829-1836.
- ↑ E. Neher and B. Sakmann."Single-channel currents recorded from membrane of denervated frog muscle fibres " Nature. 286. (1976) 71-73.
- ↑ D. T. Sawyer, "Electrochemistry for Chemists". 2nd Ed. 1995: Wiley Interscience.
- ↑ Alireza Mashaghi et al. Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics Anal. Chem., 80 (10), 3666–3676 (2008)
- ↑ M. Bonn et al., Structural inhomogeneity of interfacial water at lipid monolayers revealed by surface-specific vibrational pump-probe spectroscopy, J. Am. Chem. Soc. 132, 14971–14978 (2010).
- ↑ Mischa Bonn et al., Interfacial Water Facilitates Energy Transfer by Inducing Extended Vibrations in Membrane Lipids, J Phys Chem, 2012 http://pubs.acs.org/doi/abs/10.1021/jp302478a
- ↑ 13.0 13.1 T. Kaasgaard, C. Leidy, J. H. Crowe, O. E. Mouritsen and K. Jorgensen."Temperature-Controlled Structure and Kinetics of Ripple Phases in One- and Two-Component Supported Lipid Bilayers " Biophysical Journal. 85. (2003) 350-360.
- ↑ Y. Roiter, M. Ornatska, A. R. Rammohan, J. Balakrishnan, D. R. Heine, and S. Minko, Interaction of Nanoparticles with Lipid Membrane, Nano Letters, vol. 8, iss. 3, pp. 941-944 (2008).
- ↑ R. P. Richter and A. Brisson."Characterization of lipid bilayers and protein assemblies supported on rough surfaces by atomic force microscopy." Langmuir. 19. (2003) 1632-1640.
- ↑ S. Steltenkamp, M. M. Muller, M. Deserno, C. Hennesthal, C. Steinem and A. Janshoff."Mechanical properties of pore-spanning lipid bilayers probed by atomic force microscopy." Biophysical Journal. 91. (2006) 217-226.
- ↑ S. W. Hui, R. Viswanathan, J. A. Zasadzinski and J. N. Israelachvili."The structure and stability of phospholipid bilayers by atomic force microscopy." Biophysical Journal. 68. (1995) 171-8.
- ↑ K. Dimitrievski, M. Zach, V. P. Zhadanov and B. Kasemo."Imaging and manipulation of adsorbed lipid vesicles by an AFM tip : Experiment and Monte Carlo simulations." Colloids and Surfaces B. 47. (2006) 115-125.
- ↑ J. Schneider, W. Barger and G. U. Lee."Nanometer scale surface properties of supported lipid bilayers measured with hydrophobic and hydrophilic atomic force microscope probes." Langmuir. 19. (2003) 1899-1907.
- ↑ J. D. Robertson."The molecular structure and contact relationships of cell membranes." Progress Biophysics and Biophysical Chemistry. 10. (1960) 343-418.
- ↑ J. E. Heuser, T. S. Reese, M. J. Dennis, Y. Jan, L. Jan and L. Evans."Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release." Journal of Cell Biology. 81. (1979) 275-300.
- ↑ D. Papahadjapoulos and N. Miller."Phospholipid Model Membranes I. Structural characteristics of hydrated liquid crystals." Biochimica et Biophysica Acta. 135. (1967) 624-638.
- ↑ D. M. Small."Phase equilibria and structure of dry and hydrated egg lecithin " Journal of Lipid Research. 8. (1967) 551-557.
- ↑ G. Zaccai, J. K. Blasie and B. P. Schoenborn."Neutron diffraction studies on the location of water in lecithin bilayer model membranes." Proceedings of the National Academy of Sciences of the United States of America. 72. (1975) 376-380.
- ↑ D. Boal, "Mechanics of the Cell". 2002, Cambridge, UK: Cambridge University Press.
 |