Biology:MIAT (gene)
 Generic protein structure example |
MIAT (myocardial infarction-associated transcript), also known as RNCR2 (retinal non-coding RNA 2) or Gomafu, is a long non-coding RNA. Single nucleotide polymorphisms (SNPs) in MIAT are associated with a risk of myocardial infarction.[1] It is expressed in neurons, and located in the nucleus.[2] It plays a role in the regulation of retinal cell fate specification.[3] Crea and collaborators have shown that MIAT is highly up-regulated in aggressive prostate cancer samples,[4] raising the possibility that this gene plays a role in cancer progression.[5]
Structure
The MIAT gene is located on Chromosome 22 and is 30,051 bases in length.[6] MIAT's other name, gomafu, is a word in Japanese that means “spotted pattern”.[7] The reason it is named as so is because Gomafu is distributed in the nucleoplasm in a spotted pattern.[7] Moreover, its orientation is a plus strand.[6]
It is also found that MIAT has five exons and is likely to be a functional RNA, since MIAT hasn’t been shown to encode any translational product.[6] Furthermore, the gene encodes a spliced, long non-coding RNA.[6]
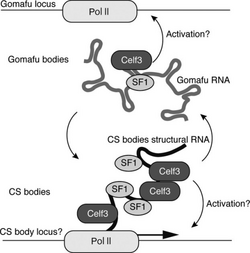
The gene is found not only in humans, but also in mice and rats.[7] Orthologs are present in syntenic positions of frog and chicken.[8] It is also found that all gomafu RNA contain tandem repeats of UACUAAC that binds to SPF1, which is a splicing factor.[7]
MIAT was originally discovered as long intergenic noncoding RNAs quite enriched in specific neurons in mouse retina and later more widely expressed in the nervous system and cultured neurons, where it specifies cell identity.[7] Moreover, the gomafu RNA is also quite insoluble[7] and is enriched in PolyA+. Also, there are putative polyadenylation signals (ATTAAA) found at the 3’ end of this gene. The presence of a PolyA tail and multiple exons and introns fulfills the feature of mRNAs transcribed by RNA polymerase II. The stability of the gene is not significantly different from β-actin mRNA.[7]
Function in Pathology
Myocardial Infarction
Myocardial infarction is more commonly known as a heart attack. It is the irreversible death of the heart muscle due to prolonged obstruction of blood supply to the organ. Case-controlled large scale studies utilizing Single Nucleotide Polymorphisms(SNPs) throughout the genome demonstrated that altered expression at 6 SNPs in the MIAT gene might confer genetic susceptibility to myocardial infarctions. MIAT has been demonstrated to encode a nonfunctional RNA. Although the exact function of MIAT is still unclear, knowledge of some of the genetic factors that contribute to the pathogenesis of myocardial infarction can lend itself to better diagnosis, prevention, and treatment. Despite all that has been discovered about MIAT, a causal link between MIAT and myocardial infarctions has not yet been demonstrated.[6]
Additionally, one study demonstrated that expression levels of MIAT are shown to change in peripheral blood cells of patients with acute myocardial infarction. In particular, researchers studied the association between levels of lncRNAs and inflammation markers in patients who have suffered a myocardial infarction. MIAT levels were found to be positively associated with lymphocytes and negatively associated with neutrophils and platelets. In another portion of this study, researchers looked at the association between cardiovascular risk factors and levels of lncRNAs. Smoking was a cardiovascular risk factor that was found to be positively associated with MIAT. It is important to realize that even though several researchers have reported that levels of lncRNAs are regulated in the cardiac tissue following a heart attack, it is not known for sure whether it is the myocardial infarction that affects the levels of lncRNAs in peripheral blood cells.[4] MIAT has various genotypes of SNPs and it is possible that only one of them relates to heart disease.[9]
Schizophrenia
The long non-coding RNA(lncRNA) MIAT is located in the same chromosomal region which is linked to Schizophrenia (SZ) 22Q12.1.[10]
MIAT is upregulated in the nucleus accumbens of cocaine and heroin users.[11][12] The nucleus accumbens is a region involved in behavior and addiction,[13] suggesting that dysregulation of MIAT can influence behavior.
It is well accepted that alternative splicing has a role in SZ pathology.[14] MIAT is associated with alternative splicing through its interaction with splicing factor 1(SF1)[8] and with genes DISC1 and ERBB4.[15] MIAT binds directly to the splicing regulator quaking homolog (QKI) and serine/arginine-rich splicing factor 1 (SRSF1).[16] QKI gene expression is decreased in specific brain regions in SZ [17][18] and it has been proposed to be involved in SZ.[19][20]
Post -mortem SZ brains have upregulated expression of both DISC1 and ERBB4.[16] Overexpression of MIAT in human-induced pluripotent stem cell (HiPSC)-derived neurons shows a significant decrease in expression of both DISC1 and ERBB4 and their alternative spliced variants.[16] This is opposite to the upregulated expression seen in SZ patient brains.[16] ASO mediated knockdown of MIAT in (HiPSC)-derived neurons increase the expression of both DISC1 and ERBB4 splice variants, but not their unspliced transcripts.[16] This is almost exactly matching the aberrant splicing pattern seen in post-mortem SZ patient’s brains.[16] These results suggests that loss of function mutations or decreased expression of MIAT is involved in driving aberrant cortical splicing patterns observed in SZ post-mortem brains.[16]
Other Pathologies
MIAT up-regulation and down-regulation has been linked to various types of cancer and other pathologies.
In a study of glioblastoma multiforme, increased expression of MIAT was linked to increased survival rates.[21] In addition, the glioma cells were found to how significantly down-regulated MIAT.[21] The role of MIAT in lymphocytic leukemia is very different from that of glioblastoma. In certain aggressive cell lines of chronic lymphocytic leukemias, MIAT is upregulated and depends on the presence of a transcriptional regulator, OCT4.[22] OCT4 serves a positive regulator of MIAT transcription and as of now is the only known regulator. However, analysis of relative concentrations of MIAT and OCT4 have indicated that other regulatory factors are in play.[22]
Beyond its role in cancer, MIAT misexpression has also been linked to neurovascular dysfunction.[23]
References
- ↑ "Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction". Journal of Human Genetics 51 (12): 1087–1099. 2006. doi:10.1007/s10038-006-0070-9. PMID 17066261.
- ↑ "The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons". Journal of Cell Science 120 (Pt 15): 2498–2506. August 2007. doi:10.1242/jcs.009357. PMID 17623775.
- ↑ "The long noncoding RNA RNCR2 directs mouse retinal cell specification". BMC Developmental Biology 10: 49. May 2010. doi:10.1186/1471-213X-10-49. PMID 20459797.
- ↑ 4.0 4.1 "Long noncoding RNAs in patients with acute myocardial infarction" (in en). Circulation Research 115 (7): 668–677. September 2014. doi:10.1161/CIRCRESAHA.115.303836. PMID 25035150.
- ↑ "The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer". Epigenomics 8 (5): 721–731. 2016. doi:10.2217/epi.16.6. PMID 27096814. http://oro.open.ac.uk/46262/4/Crea_ORO.pdf.
- ↑ 6.0 6.1 6.2 6.3 6.4 "Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction" (in en). Journal of Human Genetics 51 (12): 1087–1099. 2006-10-26. doi:10.1007/s10038-006-0070-9. PMID 17066261.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 "The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons" (in en). Journal of Cell Science 120 (Pt 15): 2498–2506. August 2007. doi:10.1242/jcs.009357. PMID 17623775.
- ↑ 8.0 8.1 "Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1" (in en). Genes to Cells 16 (5): 479–490. May 2011. doi:10.1111/j.1365-2443.2011.01502.x. PMID 21463453.
- ↑ "Variation of gene-based SNPs and linkage disequilibrium patterns in the human genome" (in en). Human Molecular Genetics 13 (15): 1623–1632. August 2004. doi:10.1093/hmg/ddh177. PMID 15190013.
- ↑ Takahashi, Sakae; Ohtsuki, Tsuyuka; Yu, Shun-Ying; Tanabe, Ei-Ichi; Yara, Kazuo; Kamioka, Masashi; Matsushima, Eisuke; Matsuura, Masato et al. (2003). "Significant linkage to chromosome 22q for exploratory eye movement dysfunction in schizophrenia". American Journal of Medical Genetics 123B (1): 27–32. doi:10.1002/ajmg.b.10046. PMID 14582142.
- ↑ "Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse" (in en). Neuropsychopharmacology 31 (10): 2304–2312. October 2006. doi:10.1038/sj.npp.1301089. PMID 16710320.
- ↑ "Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers" (in en). Journal of Neurochemistry 116 (3): 459–466. February 2011. doi:10.1111/j.1471-4159.2010.07126.x. PMID 21128942.
- ↑ "Dopamine and drug addiction: the nucleus accumbens shell connection". Neuropharmacology. Frontiers in Addiction Research: Celebrating the 30th Anniversary of the National Institute on Drug Abuse. 47 (Suppl 1): 227–241. 2004-01-01. doi:10.1016/j.neuropharm.2004.06.032. PMID 15464140.
- ↑ "Aberrant regulation of alternative pre-mRNA splicing in schizophrenia". Neurochemistry International 57 (7): 691–704. December 2010. doi:10.1016/j.neuint.2010.08.012. PMID 20813145.
- ↑ "Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia" (in en). Human Molecular Genetics 16 (2): 129–141. January 2007. doi:10.1093/hmg/ddl449. PMID 17164265.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 "The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing". Molecular Psychiatry 19 (4): 486–494. April 2014. doi:10.1038/mp.2013.45. PMID 23628989.
- ↑ "The human homolog of the QKI gene affected in the severe dysmyelination "quaking" mouse phenotype: downregulated in multiple brain regions in schizophrenia". The American Journal of Psychiatry 163 (10): 1834–1837. October 2006. doi:10.1176/ajp.2006.163.10.1834. PMID 17012699.
- ↑ "Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia". Schizophrenia Research 90 (1–3): 15–27. February 2007. doi:10.1016/j.schres.2006.11.017. PMID 17223013.
- ↑ "Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia" (in en). Proceedings of the National Academy of Sciences of the United States of America 103 (19): 7482–7487. May 2006. doi:10.1073/pnas.0601213103. PMID 16641098. Bibcode: 2006PNAS..103.7482A.
- ↑ "Human QKI, a new candidate gene for schizophrenia involved in myelination" (in en). American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics 141B (1): 84–90. January 2006. doi:10.1002/ajmg.b.30243. PMID 16342280.
- ↑ 21.0 21.1 "A long non-coding RNA signature in glioblastoma multiforme predicts survival". Neurobiology of Disease 58: 123–131. October 2013. doi:10.1016/j.nbd.2013.05.011. PMID 23726844.
- ↑ 22.0 22.1 "Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias". Oncotarget 7 (34): 54174–54182. August 2016. doi:10.18632/oncotarget.11099. PMID 27527866.
- ↑ "Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain". Oncotarget 7 (31): 49688–49698. July 2016. doi:10.18632/oncotarget.10434. PMID 27391072.
 |