Biology:Major capsid protein VP1
| Major capsid protein VP1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
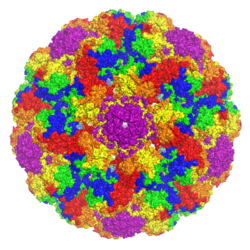
 A rendering of an icosahedral viral capsid comprising 72 pentamers of murine polyomavirus VP1, colored such that areas of the surface closer to the interior center appear blue and areas further away appear red. Rendered from PDB: 1SIE. | |||||||||
| Identifiers | |||||||||
| Symbol | VP1 | ||||||||
| Pfam | PF00718 | ||||||||
| InterPro | IPR000662 | ||||||||
| |||||||||
Major capsid protein VP1 is a viral protein that is the main component of the polyomavirus capsid. VP1 monomers are generally around 350 amino acids long and are capable of self-assembly into an icosahedral structure consisting of 360 VP1 molecules organized into 72 pentamers. VP1 molecules possess a surface binding site that interacts with sialic acids attached to glycans, including some gangliosides, on the surfaces of cells to initiate the process of viral infection. The VP1 protein, along with capsid components VP2 and VP3, is expressed from the "late region" of the circular viral genome.[1][2][3]
Structure
VP1 is the major structural component of the polyomavirus icosahedral capsid, which has T=7 symmetry and a diameter of 40-45 nm. The capsid contains three proteins; VP1 is the primary component and forms a 360-unit outer capsid layer composed of 72 pentamers. The other two components, VP2 and VP3, have high sequence similarity to each other, with VP3 truncated at the N-terminus relative to VP2. VP2 and VP3 assemble inside the capsid in contact with VP1,[1][2] with a stoichiometry of one VP2 or VP3 molecule to each pentamer.[4][5]:314 VP1 is capable of self-assembly into virus-like particles even in the absence of other viral components.[6] This process requires bound calcium ions and the resulting particles are stabilized by, but do not require, inter-pentamer disulfide bonds.[7]
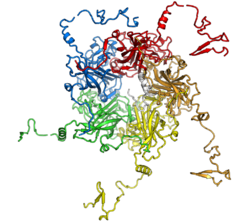
The VP1 protein monomer is primarily composed of beta sheets folded into a jelly roll fold. Interactions between VP1 molecules within a pentamer involve extensive binding surfaces, mediated in part by interactions between edge beta-strands. The VP1 C-terminus is disordered and forms interactions between neighboring pentamers in the assembled capsid. The flexibility of the C-terminal arm will enable it to adopt different conformations in the six distinct interaction environments imposed by the symmetry of the icosahedral assembly.[4][8] The C-terminus also contains a basic nuclear localization sequence,[5]:316 while the N-terminus - which is oriented toward the center of the assembled capsid - contains basic residues that facilitate non-sequence-specific interactions with DNA.[9]
Function and trafficking

The VP1 protein is responsible for initiating the process of infecting a cell by binding to sialic acids in glycans, including some gangliosides, on the cell surface.[3][8][10] Canonically, VP1 interacts specifically with α(2,3)-linked and α(2,6)-linked sialic acids.[3][8] In some cases additional factors are necessary conditions for viral entry; for example, JC virus requires the 5HT2A serotonin receptor for entry, although the specific mechanism of this requirement is unclear.[11] Once attached to the cell surface, the virions enter the cell and are trafficked by a retrograde pathway to the endoplasmic reticulum. The exact mechanism of endocytosis varies depending on the virus, and some viruses use multiple mechanisms; caveolae-dependent mechanisms are common.[12] The process by which polyomaviruses penetrate the membrane and exit the ER is not well understood, but conformational changes to VP1, possibly including reduction of its disulfide bonds, likely occur in the ER. For some polyomaviruses, VP1 has been detected reaching the nucleus along with the viral genome, though it is unclear how the genomic DNA disengages from VP1.[12]
All of the capsid proteins are expressed from the late region of the viral genome, so named because expression occurs only late in the infection process. VP1 has a nuclear localization sequence that enables import from the cytoplasm where it is synthesized by the host translation machinery to the cell nucleus where new virions are assembled. This nuclear import process, mediated by karyopherins, acts on assembled VP1 pentamers in complex with VP2 or VP3; oligomerization to form capsids occurs in the nucleus.[5]:316–17
References
- ↑ 1.0 1.1 "Murine polyomavirus tumour specific transplantation antigens and viral persistence in relation to the immune response, and tumour development". Seminars in Cancer Biology 19 (4): 236–43. August 2009. doi:10.1016/j.semcancer.2009.02.001. PMID 19505651.
- ↑ 2.0 2.1 "Lessons from immune responses and vaccines against murine polyomavirus infection and polyomavirus-induced tumours potentially useful for studies on human polyomaviruses". Anticancer Research 30 (2): 279–84. February 2010. PMID 20332429.
- ↑ 3.0 3.1 3.2 3.3 "Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity". PLoS Pathogens 11 (10): e1005104. October 2015. doi:10.1371/journal.ppat.1005104. PMID 26474293.
- ↑ 4.0 4.1 "Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry". The EMBO Journal 17 (12): 3233–40. June 1998. doi:10.1093/emboj/17.12.3233. PMID 9628860.
- ↑ 5.0 5.1 5.2 Almendral, José M. (2013). "Assembly of Simple Icosahedral Viruses". in Mateu, Mauricio G.. Structure and physics of viruses an integrated textbook. Dordrecht: Springer. ISBN 978-94-007-6552-8.
- ↑ "Self-assembly of purified polyomavirus capsid protein VP1". Cell 46 (6): 895–904. September 1986. doi:10.1016/0092-8674(86)90071-1. PMID 3019556.
- ↑ "Mechanism of assembly of recombinant murine polyomavirus-like particles". Journal of Virology 74 (4): 1658–62. February 2000. doi:10.1128/jvi.74.4.1658-1662.2000. PMID 10644335.
- ↑ 8.0 8.1 8.2 "Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments". Structure 4 (2): 183–94. February 1996. doi:10.1016/s0969-2126(96)00021-4. PMID 8805524.
- ↑ "Characterization of the DNA-binding properties of the polyomavirus capsid protein VP1". Journal of Virology 65 (3): 1168–76. March 1991. PMID 1847446.
- ↑ "Gangliosides are receptors for murine polyoma virus and SV40". The EMBO Journal 22 (17): 4346–55. September 2003. doi:10.1093/emboj/cdg439. PMID 12941687.
- ↑ "JC polyomavirus attachment, entry, and trafficking: unlocking the keys to a fatal infection". Journal of Neurovirology 21 (6): 601–13. December 2015. doi:10.1007/s13365-014-0272-4. PMID 25078361.
- ↑ 12.0 12.1 "Cellular entry of polyomaviruses". Current Topics in Microbiology and Immunology 343: 177–94. 2010. doi:10.1007/82_2010_38. PMID 20373089.