Biology:Marnaviridae
| Marnaviridae | |
|---|---|

| |
| Low-resolution image of a Marnavirus under electron microscope | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Picornavirales |
| Family: | Marnaviridae |
| Genera[1] | |
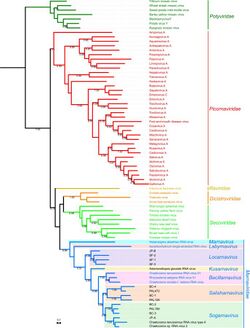 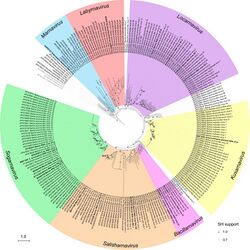 | |
Marnaviridae is a family of positive-stranded RNA viruses in the order Picornavirales that infect various photosynthetic marine protists.[2] Members of the family have non-enveloped, icosahedral capsids. Replication occurs in the cytoplasm and causes lysis of the host cell. The first species of this family that was isolated is Heterosigma akashiwo RNA virus (HaRNAV) in the genus Marnavirus,[3] which infects the toxic bloom-forming Raphidophyte alga, Heterosigma akashiwo.[4] As of 2021, there are twenty species across seven genera in this family, as well as many other related virus sequences discovered through metagenomic sequencing that are currently unclassified.[5]
Interactions between members of the Marnaviridae family and their hosts have notable significance in marine ecology, and are also relevant within the aquaculture industry. HaRNAV and viruses from Bacillarnavirus are known to have roles in regulating dynamics and composition of their hosts’ blooms.[6][7] An unclassified sequence, Baishivirus, has been suggested to be the possible pathogen of glass post-larvae disease, which is prevalent in shrimp aquaculture.[8] Viruses detected in a cultured prawn species that had been affected by growth retardation disease have also been placed in Marnaviridae.[9]
History
The name "marnaviridae" is based on its genome type (RNA virus - rnaviridae), together with the prefix "ma" being derived from the Latin word mare (sea).[10]
The family was proposed following the discovery of an RNA virus (HaRNAV) that infects H. akashiwo off of the coast of British Columbia, which was the first report of a single-stranded RNA virus capable of causing cell lysis in phytoplankton.[6]
HaRNAV was isolated from water collected in the Strait of Georgia in British Columbia, Canada, from a concentrated virus assemblage using the host H. akashiwo (NEPCC 522).[6] It must not be confused with two other unrelated viruses that infect this host, H. akashiwo virus 01 (HaV-1, isolate: HaV53) in the genus Raphidovirus, and Heterosigma akashiwo nuclear inclusion virus (HaNIV).[11]
Marnaviridae existed for multiple years with Marnavirus as the only genus and HaRNAV as the only species. After the usage of metagenomic analysis on the amino acid sequences of the capsid proteins and RdRp domains on viruses under the order Picornavirales, Marnaviridae was discovered to have a larger variety of viruses classified under it. Previously unassigned Labyrnavirus and Bacillarnavirus were also classified as genera under Marnaviridae.[10]
Further metagenomic studies have yielded 653 Marnavirus-like sequences,[12] prompting the proposal for a taxonomic reorganization to incorporate the new data into existing frameworks.[13]
Taxonomy
The following genera are recognised:[14]
- Bacillarnavirus
- Kusarnavirus
- Labyrnavirus
- Locarnavirus
- Marnavirus
- Salisharnavirus
- Sogarnavirus
Classification
While Marnaviridae has several characteristics that are similar to those of other viruses in the broader Picornavirales order, members of Marnaviridae are placed through analysis of the RdRp sequence of amino acids.[5] The seven genera of Marnaviridae are designated based on analysis of RNA-dependent RNA polymerase (RdRp) sequences and capsid amino acid sequences,[10] species are listed under each genus.[5]
Bacillarnavirus
The Bacillarnavirus species share 57.8-64.4% of RdRp and 29.1-33.4% of capsid amino acid sequences, when compared with each other within Bacillarnavirus. When compared with other genera of the family, they share 23.8-47.8% of RdRp and 18.3-29.4% of capsid sequences.[10]
- Chaetoceros socialis forma radians RNA virus 1
- Chaetoceros tenuissimus RNA virus 01
- Rhizosolenia setigera RNA virus 01
Kusarnavirus
Kusarnavirus shares 25.4-48.2% of RdRp and 21.7-47.9% of capsid sequences, with other genera in the family.
- Astarnavirus
Labyrnavirus
Labyrnavirus shares 22.4-33.6% identities of RdRp and 17.1-22.5% of capsid sequences. It is a deep-branching taxon.[10]
- Aurantiochytrium single-strand RNA virus 01
Locarnavirus
The Locarnavirus species share 52.4-59.1% identities of RdRp and 32.2-38.3% of capsid sequences, when compared with each other within Locarnavirus. When compared with other genera of the family, they share 24.0-48.2% of RdRp and 17.2-27.4% of capsid sequences.
- Jericarnavirus B
- Sanfarnavirus 1
- Sanfarnavirus 2
- Sanfarnavirus 3
Marnavirus
Marnavirus shares 22.4-30.3% of amino acid sequence identity of RdRp and 18.5-26.4% of capsid sequences. It is the most divergent and deeply branched taxon within the family.[10]
- Heterosigma akashiwo RNA virus
Salisharnavirus
The Salisharnavirus species share 35.6-63.6% of RdRp and 24.9-34.4% of capsid sequences, when compared with each other within Salisharnavirus. When compared with other genera of the family, they share 24.3-47.2% of RdRp and 18.2-31.5% of capsid sequences.
- Britarnavirus 1
- Britarnavirus 4
- Palmarnavirus 128
- Palmarnavirus 473
Sogarnavirus
The Sogarnavirus species share 48.8-77.0% of RdRp and 27.6-59.0% of capsid sequences, when compared with each other within Sogarnavirus. When compared with other genera of the family, they share 24.3-47.2% of RdRp and 17.1-47.9% for capsid sequences.
- Britarnavirus 2
- Britarnavirus 3
- Chaetarnavirus 2
- Chaetenuissarnavirus II
- Jericarnavirus A
- Palmarnavirus 156
Additionally, there are 653 virus sequences that have been identified (via metagenomic analysis) as possibly belonging to the family Marnaviridae. However, these sequences are not assigned under any of the existing genera.[12]
Characteristics
Structure

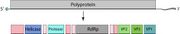
Modified after ViralZone.[15]


Virions in Marnaviridae are non-enveloped, with icosahedral geometries, and T=pseudo3 symmetry. Virus particles are 22-35 nm in diameter. The capsid consists of three major capsid proteins (MCPs: VP1, VP2, VP3), each having a Jelly roll fold. Most species also encode a minor capsid protein (mCP: VP4). All VPs are composed of β-sheets and α-helices. that is located around the five-fold axes on the inside of the capsid.[16]
The structures of Chaetoceros socialis forma radians RNA virus 1 (CsfrRNAV01, Bacillarnavirus) and Chaetoceros tenuissimus RNA virus type II (CtenRNAVII, Sogarnavirus) have been determined. In both species, VP1 surrounds the five-fold axes while VP2 and VP3 form the two- and three-fold axes. The jelly roll folds on the MCPs are non-canonical with VP1 having nine β-strands and VP2 and VP3 having seven. VP1 also contains a unique EF-loop connected to the extra β-strand in the jelly roll fold which protrudes from the surface of the virus particle. A CD-loop is also present on VP1 which obscures the canyon structure that binds host receptors in the related Picornaviridae family.[16] Unlike other species, VP4 is not present in the CsfrRNAV01 capsid, although the corresponding gene still exists.[17]
Genome
Marnaviridae genomes consist of linear, non-segmented, positive-sense single-stranded RNA 8.6-9.6 kb in length.[15] The genomic RNA generally contains either one or two open-reading frames (ORFs) that may overlap. For species whose genomes have two ORFs, ORF1 encodes proteins involved in replication (RNA helicase, RdRp) and a cysteine protease which cleaves the polyprotein, while ORF2 encodes structural proteins.[18] Genomes also contain internal ribosome entry sites (IRES) in intergenic regions and on the 5' end. The 3' end of the genome contains a poly(A) tail.[5]
One exception to the typical genome organization of Marnaviridae is Aurantiochytrium single-strand RNA virus 01 (AuRNAV01) which has a third ORF that encodes a protein with unknown function.[19] In AuRNAV01, ORFs 2 and 3 are thought to be translated from subgenomic RNA rather than using an IRES.
Host interactions
Life cycle
Various marine phytoplankton species serve as hosts for members of the family Marnaviridae. Entry of the virus is achieved by penetration into the host cell. EF-, CD-, and E1E2-loops are hypothesized to interact with host receptors to trigger viral entry.[17]
Replication occurs in the cytoplasm and relies on the viral RdRp.[14] The positive-sense RNA genome is directly translated to produce viral proteins.[14] After a latent period between 8–48 hours, host cells are observed to experience cytopathic symptoms such as swelling of the endoplasmic reticulum, formation of vacuoles, appearance of fibrous matter in vacuolated parts of the cell, and the deterioration of the cytoplasm.[14] Virus particles accumulate in the cytoplasm and may form crystalline arrays or random aggregates.[18] The virus then exits the host cell by tubule-guided viral movement and virions are released by cell lysis.[15] Burst sizes differ greatly between species and range from 66 to 106.[14] As infected cells are lysed shortly after showing signs of infection, proportions of infected cells in the environment remain low.[20]
Marnavirus
Heterosigma akashiwo RNA virus (HaRNAV)
HaRNAV is the first identified virus in the Marnaviridae family.[4] When the virus infects H. akashiwo, a toxic bloom-forming alga, it replicates inside the cytoplasm of its host, and is distributed either singly or in groups as crystalline arrays. Within those that are aligned in crystalline arrays, sectioned particles show that they are either packaged completely (electron dense), or incompletely (ring-shaped). Once infected, the host cells exhibit symptoms such as swelling of the endoplasmic reticulum, disintegration and vacuolation of the cytoplasm, as well as fibrous material appearing in those vacuolated areas.
HaRNAV is known to be able to infect multiple strains of H. akashiwo in the Pacific Ocean, namely 3 strains (NEPCC 102, NEPCC 522, NEPCC 764) from the Northeast Pacific region, and 2 strains (H93616, H94608) from Japan. It is noted that HaRNAV infect different strains of H. akashiwo than other H. akashiwo viruses, suggesting co-existence, with each virus having a slightly different host range. H. akashiwo cells also demonstrate more viral resistance after its bloom has collapsed, suggesting that the H. akashiwo strains may change over the course of the algal bloom, which is regulated by the viruses.[6]
Bacillarnavirus
Chaetoceros tenuissimus RNA virus 01
CtenRNAV01 infects the bloom-forming diatom Chaetoceros tenussimus Meunier. It was first isolated from strain 2-10 but is able to infect other strains of the same species. Host cells in stationary phase are more susceptible to the virus than logarithmic phase cells.[18] Host susceptibility is also affected by temperature, with various strains being more susceptible at temperatures above 20 °C. Strain 2–10, however, is more susceptible below 20 °C which is not optimal for host growth.[21] Iron-limitation has also been shown to reduce viral infection rates.[22] Stationary phase infection and virus-induced lysis of host cells is also impaired by bacteria of the genera Nautella, Polaribacter, and Sulitobacter that coexist with the host diatom in the marine environment.[23] This is thought to be a mechanism allowing for susceptible diatom species to survive viral infections.
Chaetoceros socialis forma radians RNA virus 1
The diatom C. socialis is the natural host of CsfrRNAV01. Infection with the virus induces spore formation in host cells, and the spores formed take longer to germinate than spores induced by host nutrient limitation. Although viral RNA is present within these spores, those that germinate are not able to produce infectious viral particles.[24]
Sogarnavirus
Chaetoceros tenuissimus RNA virus type II
Like CtenRNAV01, CtenRNAVII was first isolated from C. tenussimus Meunier strain 2–10, however it is also able to infect at least 4 other Chaetoceros species.[25] It is the only Marnaviridae species identified thus far that has demonstrated this broad of a host range.
Unassigned viruses
Given the nature of metaviromic analysis, it is difficult to determine the exact host range of these viruses. However, since the species fall under Marnaviridae, as well as by comparing genetic codes in other virus groups in the Yangshan assemblage, it has been inferenced that these viruses likely infect unicellular eukaryotes.[12]
Echinodermata
Through a study aiming to investigate the diversity of the RNA viruses infecting species within the phylum Echinodermata, the transcriptomes and viral metagenomes of 38 echinoderm species, with representation of species from all five classes of echinoderms, were analyzed and categorized into their respective families and genera. From this analysis, it was found that of the viral contigs recovered, more than half were classified within the viral order Picornavirales, with the highest number of the picornaviruses being placed in the Marnaviridae family.[26] This finding suggests the protists, such as symbionts, that are affiliated with echinoderms are infected by Marnaviridae, or that members of this viral family are capable of infecting a wider range of hosts than the single-celled eukaryotes they were initially known to infect.
Host-Specific Receptor-Binding Mechanisms
Marnaviridae use a narrow range of hosts, as they lyse their host species, which is thought to connect to a receptor binding mechanism unique to the family.[17] In tests involving the species Chaetoceros socialis forma radians RNA virus (CsfrRNAV), full and empty capsids were analyzed by their atomic structure to which identified common and diverse structural features of the VP1 protein surface between different virus species under Marnaviridae.[17]
Unlike viruses under Picornaviridae, Marnaviridae have an extra EF-Loop, implying the usage of a unique receptor-binding mechanism. A possible binding site for algal hosts were also found in E1E2 and/or CD loops, which could play a critical role in its unique receptor-binding mechanism.[17]
In the structural analysis of the capsid's cryo-EM maps and atomic models, PyMOL 1.4 was used to obtain a Root-Mean-Square-Deviated (RMSD) per residue, which provided a value for structural diversity. Analysis for the structural proteins VP1, VP2, and VP3, suggested that VP1 was responsible for the host-specificity of the virus, and was based on the diversity in the VP1 protein.[17]
When looking at a structural phylogeny of Marnaviridae based on VP1s, local structural diversity that is reflected can be used to better predict the targeted algal hosts of different Marnaviridae viruses.[17]
Ecology
Distribution
While HaRNAV was initially isolated from British Columbian waters, it has been found that HaRNAV is also capable of infecting H. akashiwo strains originating from Japan, thus suggesting that members of Marnaviridae may have a wide distribution throughout the Pacific Ocean.[27] As viruses within the Marnaviridae family have been identified from shrimp and prawn aquaculture species in Chinese waters, this further supports Marnaviridae being widespread across the Pacific Ocean from North America to Asia.[8][9] Furthermore, sequencing of RNA viromes originating from Limnopolar Lake in the Antarctic revealed four Antarctia picorna-like viruses (APLV), one of which, APLV-3, was found to cluster with the Marnaviridae family, thereby further expanding the family's potential distribution southwards to a cold, freshwater environment in Antarctica.[28]
Heterosigma akashiwo RNA virus (HaRNAV)
The complex relationship between HaRNAV and other viruses capable of infecting H. akashiwo is important in terms of understanding how viruses play a role in H. akashiwo bloom dynamics.[6] When compared to Heterosigma akashiwo nuclear inclusion virus (HaNIV), it is demonstrated that although both viruses infect host strains from the Northeast Pacific (with HaRNAV also able to infect host strains from Japan), they each targeted different subsets of host strains. The difference in host range between the two viruses suggests that they likely coexisted within the local environment, each infecting their specific hosts, therefore it is possible to infer the strain composition of H. akashiwo population over its blooming period.[6] Studies have shown that H. akashiwo's susceptibility to viruses tends to shift over the course of the bloom, with the cells being much more resistant to viral infection after the bloom has collapsed. This suggests that viruses may have played an important role in regulating the H. akashiwo bloom.[29] Similarly, a study done on the viral assemblage composition of HaRNAV has shown that the diversity of HaRNAV is correlated with the composition of its host, therefore also suggesting that HaRNAV influences the composition of H. akashiwo.[30]
Bacillarnavirus
In terms of ecological impact and species diversity in the ocean, the genus Chaetoceros is considered to be a major taxonomic group, playing an essential role as primary producers during bloom periods. Viruses under Bacillarnavirus have been shown to target Chaetoceros in natural environments, with different species targeting specific strains of the diatom. Studies have suggested that these viruses play important roles in controlling and regulating the population of Chaetoceros, thus influencing the spring bloom and subsequently affecting the local marine ecosystems as well.[31]
Guinardia delicatula RNA virus
GdelRNAV is a Bacillarnavirus species that infects the bloom-forming diatom Guinardia delicatula present in temperate coastal waters.[32] Strains of this virus have only been successfully isolated shortly after the summer bloom of G. delicatula. This suggests that the virus may play a role in decreasing host populations at the end of the summer bloom, although other parasites of G. delicatula are also likely involved.
Aurantiochytrium RNA virus (AuRNAV)
Thraustochytrids are considered to be important decomposers in ecosystems. Aurantiochytrium RNA virus (AuRNAV) has been shown to infect thraustochytrids together with another virus, Sicyoidochytrium minutum DNA virus (SmDNAV). Studies have revealed that these two viruses maintain different fluctuation patterns from each other, with AuRNAV showing a significant increase in abundance following an increase of Aurantiochytrium sp. thraustochytrids (which decompose organic matter during the decline of H. akashiwo bloom), while SmDNAV did not demonstrate this pattern. This suggests that AuRNAV likely targets hosts that can effectively utilize dead phytoplankton cells, therefore the virus is also a vital part of the bloom dynamic.[33]
Prevalence in aquaculture
Glass post-larvae disease
Penaeus vannamei is an important shrimp aquaculture species that is cultured globally, and most notably in China. However, due to the glass post-larvae disease (GPD), a highly pathogenic disease that spreads quickly within aquaculture sites, the industry has suffered significant losses in recent years, resulting in economic instabilities. Shrimp infected with GPD show signs of slower movements and responses, along with color change in liver pancreatic tissues (full brown to light brown). In the 24 to 48 hours after infection, shrimps also exhibit swollen gill filaments, as well as the entire body becoming more transparent and brittle (hence the name of the disease). Discovered through metagenomic sequencing of a P. vannamei sample infected with GPD, Baishivirus has been suggested to be a member under the Marnaviridae family, and was found to contain three ORFs within its 9.895 kb long genome. Baishivirus is theorized to be the primary pathogen of the disease, providing a possible entryway into finding preventive measures against GPD in the shrimp industry.[8]
Growth Retardation Disease
Prevalence of growth retardation disease (GRD), which causes decreased growth rates and precocious puberty in the freshwater prawn Macrobrachium rosenbergii, has led to significant financial losses within the Chinese aquaculture industry of this prawn.[9] Generally, mature male and female prawns are 10 cm and 8 cm in body length respectively, however when impacted by GRD, the prawns exhibit early maturity, wherein males demonstrate mating behaviours at 6 cm and females possess eggs at 5 cm.[34][35] The impacts of GRD have led to decreases in prawn production of more than 50% in some cases.[36] Through metatranscriptomic sequencing of the viromes of healthy prawns and prawns with GRD, and phylogenetic analysis of the resulting RNA sequences, four marna-like viruses were placed in the Marnaviridae family. These viruses, referred to as Macrobrachium rosenbergii viruses (MRV) MRV3, MRV4, MRV5, and MRV6, have sequences which are 9242, 8887, 8548, and 9153 base pairs in length respectively.[9] MRV3, MRV5, and MRV6 were found to be novel viruses, with MRV3 being placed in the genus Labyrnavirus, and MRV5 and MRV6 belonging to the genus Sogarnavirus. In contrast, MRV4 was characterized as being a previously identified Beihai picorna-like virus in the genus Kusarnavirus.
References
- ↑ "Virus Taxonomy: 2018b Release" (in en). March 2019. https://ictv.global/taxonomy.
- ↑ "Marnaviridae - Positive Sense RNA Viruses - Positive Sense RNA Viruses (2011) - International Committee on Taxonomy of Viruses (ICTV)" (in en). https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/232/marnaviridae.[|permanent dead link|dead link}}]
- ↑ "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. https://ictv.global/taxonomy.
- ↑ 4.0 4.1 "Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo". Virology 320 (2): 206–217. March 2004. doi:10.1016/j.virol.2003.10.015. PMID 15016544.
- ↑ 5.0 5.1 5.2 5.3 "ICTV Virus Taxonomy Profile: Marnaviridae 2021". The Journal of General Virology 102 (8): 001633. August 2021. doi:10.1099/jgv.0.001633. PMID 34356002.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 "Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae)". Journal of Phycology 39 (2): 206–217. 2003. doi:10.1046/j.1529-8817.2003.01162.x.
- ↑ "First Viruses Infecting the Marine Diatom Guinardia delicatula". Frontiers in Microbiology 9: 3235. 2019. doi:10.3389/fmicb.2018.03235. PMID 30687251.
- ↑ 8.0 8.1 8.2 "A novel virus in the family Marnaviridae as a potential pathogen of Penaeus vannamei glass post-larvae disease". Virus Research 324: 199026. January 2023. doi:10.1016/j.virusres.2022.199026. PMID 36529302.
- ↑ 9.0 9.1 9.2 9.3 "Virome Analysis of Normal and Growth Retardation Disease-Affected Macrobrachium rosenbergii". Microbiology Spectrum 10 (6): e0146222. December 2022. doi:10.1128/spectrum.01462-22. PMID 36445118.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 "Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales". Virus Evolution 5 (2): vez056. July 2019. doi:10.1093/ve/vez056. PMID 31908848.
- ↑ "A novel virus (HaNIV) causes lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae)". Journal of Phycology 37 (2): 216–222. 2001. doi:10.1046/j.1529-8817.2001.037002216.x.
- ↑ 12.0 12.1 12.2 "Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome". Nature Microbiology 5 (10): 1262–1270. October 2020. doi:10.1038/s41564-020-0755-4. PMID 32690954.
- ↑ "RNA Viruses in Aquatic Unicellular Eukaryotes". Viruses 13 (3): 362. February 2021. doi:10.3390/v13030362. PMID 33668994.
- ↑ 14.0 14.1 14.2 14.3 14.4 "Family: Marnaviridae" (in en). International Committee on Taxonomy of Viruses (ICTV). https://ictv.global/report/chapter/marnaviridae/marnaviridae.
- ↑ 15.0 15.1 15.2 15.3 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/170.html.
- ↑ 16.0 16.1 "Capsid Structure of a Marine Algal Virus of the Order Picornavirales". Journal of Virology 94 (9). April 2020. doi:10.1128/JVI.01855-19. PMID 32024776.
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 17.6 "Structural Insights into Common and Host-Specific Receptor-Binding Mechanisms in Algal Picorna-like Viruses". Viruses 14 (11): 2369. October 2022. doi:10.3390/v14112369. PMID 36366467.
- ↑ 18.0 18.1 18.2 "Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier". Applied and Environmental Microbiology 74 (13): 4022–4027. July 2008. doi:10.1128/AEM.00509-08. PMID 18469125. Bibcode: 2008ApEnM..74.4022S.
- ↑ "Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp". The Journal of General Virology 87 (Pt 3): 723–733. March 2006. doi:10.1099/vir.0.81204-0. PMID 16476996.
- ↑ "Virus-specific responses of Heterosigma akashiwo to infection". Applied and Environmental Microbiology 72 (12): 7829–7834. December 2006. doi:10.1128/AEM.01207-06. PMID 17041155. Bibcode: 2006ApEnM..72.7829L.
- ↑ "Temperature alters algicidal activity of DNA and RNA viruses infecting Chaetoceros tenuissimus" (in en). Aquatic Microbial Ecology 73 (2): 171–183. 2014-10-02. doi:10.3354/ame01713. ISSN 0948-3055. http://www.int-res.com/abstracts/ame/v73/n2/p171-183/.
- ↑ "Impaired viral infection and reduced mortality of diatoms in iron-limited oceanic regions" (in en). Nature Geoscience 14 (4): 231–237. 2021. doi:10.1038/s41561-021-00711-6. ISSN 1752-0908. Bibcode: 2021NatGe..14..231K. https://cdr.lib.unc.edu/downloads/nc580x47h.
- ↑ "Coculture with marine bacteria confers resistance to complete viral lysis of diatom cultures" (in en). Aquatic Microbial Ecology 73 (1): 69–80. 2014-08-29. doi:10.3354/ame01705. ISSN 0948-3055.
- ↑ "Virus-induced spore formation as a defense mechanism in marine diatoms". The New Phytologist 229 (4): 2251–2259. February 2021. doi:10.1111/nph.16951. PMID 32978816.
- ↑ "Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom". Applied and Environmental Microbiology 81 (3): 1120–1131. February 2015. doi:10.1128/AEM.02380-14. PMID 25452289. Bibcode: 2015ApEnM..81.1120K.
- ↑ "The RNA virome of echinoderms". The Journal of General Virology 103 (6): 001772. June 2022. doi:10.1099/jgv.0.001772. PMID 35766975.
- ↑ "Diversity of Viruses Infecting Eukaryotic Algae". Current Issues in Molecular Biology 39: 29–62. 2020. doi:10.21775/cimb.039.029. PMID 32073403.
- ↑ "Viruses of Polar Aquatic Environments". Viruses 11 (2): 189. February 2019. doi:10.3390/v11020189. PMID 30813316.
- ↑ "Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo". Applied and Environmental Microbiology 66 (11): 4916–4920. November 2000. doi:10.1128/AEM.66.11.4916-4920.2000. PMID 11055943.
- ↑ "Role of Phylogenetic Structure in the Dynamics of Coastal Viral Assemblages". Applied and Environmental Microbiology 87 (11): e02704–20. May 2021. doi:10.1128/AEM.02704-20. PMID 33741635. Bibcode: 2021ApEnM..87E2704G.
- ↑ "Isolation and characterization of a single-stranded RNA virus that infects the marine planktonic diatom Chaetoceros sp. (SS08-C03): New ssRNA virus infecting Chaetoceros" (in en). Phycological Research 61 (1): 27–36. January 2013. doi:10.1111/j.1440-1835.2012.00670.x. https://onlinelibrary.wiley.com/doi/10.1111/j.1440-1835.2012.00670.x.
- ↑ "First Viruses Infecting the Marine Diatom Guinardia delicatula". Frontiers in Microbiology 9: 3235. 2019. doi:10.3389/fmicb.2018.03235. PMID 30687251.
- ↑ "Ecological Dynamics of Two Distinct Viruses Infecting Marine Eukaryotic Decomposer Thraustochytrids (Labyrinthulomycetes, Stramenopiles)". PLOS ONE 10 (7): e0133395. 2015-07-23. doi:10.1371/journal.pone.0133395. PMID 26203654. Bibcode: 2015PLoSO..1033395T.
- ↑ "Transcriptome analysis of Macrobrachium rosenbergii: Identification of precocious puberty and slow-growing information". Journal of Invertebrate Pathology 190: 107752. May 2022. doi:10.1016/j.jip.2022.107752. PMID 35367462.
- ↑ "Investigation of the causes of the "iron shell" phenomenon of Roche marsh shrimp". Scientific Fish Farming 1: 56–58. doi:10.14184/j.cnki.issn1004-843x.2014.01.034.
- ↑ "A Novel Virus of Flaviviridae Associated with Sexual Precocity in Macrobrachium rosenbergii". mSystems 6 (3): e0000321. June 2021. doi:10.1128/mSystems.00003-21. PMID 34100644.
External links
- Viralzone: Marnaviridae
- ICTV
- NCBI: Marnaviridae (family)
- ICTV: Taxonomy History – Marnaviridae
Wikidata ☰ Q15642831 entry
 |
