Biology:Semantides
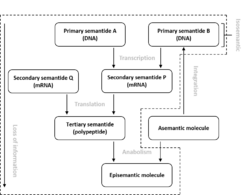
Semantides (or semantophoretic molecules) are biological macromolecules that carry genetic information or a transcript thereof. Three different categories or semantides are distinguished: primary, secondary and tertiary. Primary Semantides are genes, which consist of DNA. Secondary semantides are chains of messenger RNA, which are transcribed from DNA. Tertiary semantides are polypeptides, which are translated from messenger RNA.[1] In eukaryotic organisms, primary semantides may consist of nuclear, mitochondrial or plastid DNA.[2] Not all primary semantides ultimately form tertiary semantides. Some primary semantides are not transcribed into mRNA (non-coding DNA) and some secondary semantides are not translated into polypeptides (non-coding RNA). The complexity of semantides varies greatly. For tertiary semantides, large globular polypeptide chains are most complex while structural proteins, consisting of repeating simple sequences, are least complex. The term semantide and related terms were coined by Linus Pauling and Emile Zuckerkandl.[1] Although semantides are the major type of data used in modern phylogenetics, the term itself is not commonly used.
Related terms
Isosemantic
DNA or RNA that differs in base sequence, but translate into identical polypeptide chains are referred to as being isosemantic.[1]
Episemantic
Molecules that are synthesized by enzymes (tertiary semantides) are referred to as episemantic molecules. Episemantic molecules have a larger variety in types than semantides, which only consist of three types (DNA, RNA or polypeptides). Not all polypeptides are tertiary semantides. Some, mainly small polypeptides, can also be episemantic molecules.[1]
Asemantic
Molecules that are not produced by an organism are referred to as asemantic molecules, because they do not contain any genetic information. Asementic molecules may be changed into episemantic molecules by anabolic processes. Asemantic molecules may also become semantic molecules when they integrate into a genome. Certain viruses and episomes have this ability.[1]
When referring to a molecule as being semantic, episemantic or asemantic, then this only applies to a specific organism. A semantic molecule for one organism may be asemantic for another organism.
Research applications
Semantides are used as phylogenetic information for studying the evolutionary history of organisms. Primary semantides are also used in comparative biodiversity analyses. However, since extracellular DNA can persist for some time, these types of analysis cannot discern active from inactive and or dead organisms.[3][4]
The extent to which biological macromolecules are informative for studying evolutionary history differs. The more complex a molecule, the more informative it is in for phylogenetics. Primary and secondary semantides contain the most information. In tertiary semantides, some information is lost, because many amino acids are coded for by more than one codon.[1][5]
Episemantic molecules (e.g. carotenoids) are also informative for phylogenetics. However, the distributions of these molecules do not correlate perfectly with phylogenies based on semantides.[6] Therefore, independent confirmation is often still needed.[1] The more enzymes involved in a synthesis pathway, the more unlikely that such pathways have evolved separately. Therefore, for episemantic molecules, molecules that are synthesized from the least complex asemantic molecules are the most informative in phylogenetics. However, different pathways may synthesize similar or even identical molecules. For example, in animals, plants and other eukaryotes, different pathways have been found for vitamin C synthesis.[7] Therefore, certain molecules should not be used for studying phylogenetic relationships.[1]
Although asemantic molecules could indicate some quantitative or qualitative features of a group of organisms, they are considered to be unreliable and uninformative for phylogenetics.[1]
Analyses using different semantides may yield conflicting phylogenies. However, if the phylogenies are congruent, then there is more support for the evolutionary relationship. By analyzing larger sequences (e.g. complete mitochondrial genome sequences), phylogenies can be constructed, which are more resolved and have more support.[8]
Examples
Semantides often used in studies are common to most organisms and are known to only change slowly over time. Examples of these macromolecules are:
- ATPase
- Cytochrome b[9][10]
- Cytochrome c oxidase subunit I[2][11]
- Heat shock protein genes
- Histone H3[11]
- RecA[12]
- Recombination activating gene 1[10]
- Ribonuclease P RNA[13]
- Ribosomal DNA (e.g. 28S rDNA)[14]
- Ribosomal RNA (e.g. 16S rRNA)[2][11][12][13][15]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Zuckerkandl, Emile; Pauling, Linus (1965-03-01). "Molecules as documents of evolutionary history". Journal of Theoretical Biology 8 (2): 357–366. doi:10.1016/0022-5193(65)90083-4. ISSN 0022-5193. PMID 5876245.
- ↑ 2.0 2.1 2.2 Fontanilla, Ian Kendrich; Naggs, Fred; Wade, Christopher Mark (September 2017). "Molecular phylogeny of the Achatinoidea (Mollusca: Gastropoda)". Molecular Phylogenetics and Evolution 114: 382–385. doi:10.1016/j.ympev.2017.06.014. PMID 28647619. http://eprints.nottingham.ac.uk/44214/1/Manuscript%20final_foropenaccess.pdf.
- ↑ England, L.S; Vincent, M.L; Trevors, J.T; Holmes, S.B (2004-10-18). "Extraction, detection and persistence of extracellular DNA in forest litter microcosms". Molecular and Cellular Probes 18 (5): 313–319. doi:10.1016/j.mcp.2004.05.001.
- ↑ Rettedal, Elizabeth A.; Brözel, Volker S. (April 2015). "Characterizing the diversity of active bacteria in soil by comprehensive stable isotope probing of DNA and RNA with H 2 18 O". MicrobiologyOpen 4 (2): 208–219. doi:10.1002/mbo3.230. PMID 25650291.
- ↑ Weisblum, B.; Benzer, S.; Holley, R. W. (1962-08-01). "A Physical Basis for Degeneracy in the Amino Acid Code". Proceedings of the National Academy of Sciences 48 (8): 1449–1454. doi:10.1073/pnas.48.8.1449. ISSN 0027-8424. PMID 14005813.
- ↑ Klassen, J. L.; Foght, J. M. (2008-04-01). "Differences in Carotenoid Composition among Hymenobacter and Related Strains Support a Tree-Like Model of Carotenoid Evolution". Applied and Environmental Microbiology 74 (7): 2016–2022. doi:10.1128/AEM.02306-07. ISSN 0099-2240. PMID 18263749.
- ↑ Wheeler, Glen; Ishikawa, Takahiro; Pornsaksit, Varissa; Smirnoff, Nicholas (2015-03-13). "Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes". eLife 4. doi:10.7554/eLife.06369. ISSN 2050-084X. PMID 25768426.
- ↑ Powell, Alexis F.L.A.; Barker, F. Keith; Lanyon, Scott M. (January 2013). "Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: An avian case study". Molecular Phylogenetics and Evolution 66 (1): 69–79. doi:10.1016/j.ympev.2012.09.006. PMID 23000817.
- ↑ Castresana, Jose (2001-04-01). "Cytochrome b Phylogeny and the Taxonomy of Great Apes and Mammals". Molecular Biology and Evolution 18 (4): 465–471. doi:10.1093/oxfordjournals.molbev.a003825. ISSN 1537-1719. PMID 11264397.
- ↑ 10.0 10.1 Perdices, Anabel; Bohlen, Joerg; Šlechtová, Vendula; Doadrio, Ignacio (2016-01-04). Peng, Zuogang. ed. "Molecular Evidence for Multiple Origins of the European Spined Loaches (Teleostei, Cobitidae)". PLOS One 11 (1): e0144628. doi:10.1371/journal.pone.0144628. ISSN 1932-6203. PMID 26727121.
- ↑ 11.0 11.1 11.2 Gosliner, Terrence M.; Schepetov, Dimitry; Chichvarkhin, Anton; Ekimova, Irina; Carmona, Leila; Cella, Kristen (2016-12-15). "A Radical Solution: The Phylogeny of the Nudibranch Family Fionidae". PLOS One 11 (12): e0167800. doi:10.1371/journal.pone.0167800. ISSN 1932-6203. PMID 27977703.
- ↑ 12.0 12.1 Kalita, Michał; Małek, Wanda (December 2017). "Molecular phylogeny of Bradyrhizobium bacteria isolated from root nodules of tribe Genisteae plants growing in southeast Poland". Systematic and Applied Microbiology 40 (8): 482–491. doi:10.1016/j.syapm.2017.09.001. PMID 29102065.
- ↑ 13.0 13.1 Peters, I. R.; Helps, C. R.; McAuliffe, L.; Neimark, H.; Lappin, M. R.; Gruffydd-Jones, T. J.; Day, M. J.; Hoelzle, L. E. et al. (2008-05-01). "RNase P RNA Gene (rnpB) Phylogeny of Hemoplasmas and Other Mycoplasma Species". Journal of Clinical Microbiology 46 (5): 1873–1877. doi:10.1128/JCM.01859-07. ISSN 0095-1137. PMID 18337389.
- ↑ Mckenna, Duane D.; Farrell, Brian D.; Caterino, Michael S.; Farnum, Charles W.; Hawks, David C.; Maddison, David R.; Seago, Ainsley E.; Short, Andrew E. Z. et al. (2015). "Phylogeny and evolution of Staphyliniformia and Scarabaeiformia: forest litter as a stepping stone for diversification of nonphytophagous beetles". Systematic Entomology 40 (1): 35–60. doi:10.1111/syen.12093. ISSN 1365-3113.
- ↑ Naggs, Fred; Mordan, Peter B.; Wade, Christopher M. (2006-04-01). "Evolutionary relationships among the Pulmonate land snails and slugs (Pulmonata, Stylommatophora)". Biological Journal of the Linnean Society 87 (4): 593–610. doi:10.1111/j.1095-8312.2006.00596.x. ISSN 0024-4066. https://academic.oup.com/biolinnean/article/87/4/593/2691599.
